Аннотация
Prokaryotic and eukaryotic microorganisms cause spoilage of produced dairy and fat-and-oil products. In addition, these products can be contaminated with pathogenic microorganisms. The standard practice of detecting bacterial pathogens is based on the cultivation of microorganisms due to which the analysis lasts from 5 to 7 days. Molecular genetic methods can reduce the analysis time to 1-2 days. In this paper, the ready-made commercial products of the dairy and fat-and-oil industry have been analyzed for the microbiological composition using classical DNA barcoding and DNA metabarcoding. During the study, representatives of the genera Pseudomonas , Bacillus , Lactococcus , Kocuria , Staphylococcus , Moraxella , Paucisalibacillus, Acinetobacter , Klebsiella , Paenibacillus , Lysinibacillus , Enterobacter, Acetobacter and Massilia have been defined . When analyzing the quantitative ratio of microorganisms, it was revealed that dairy and fat-and-oil products are most often seeded with Bacillus sp., among which Bacillus licheniformis (16.67% of colonies) and Bacillus subtilis (11.4% of colonies) can be distinguished . Among Pseudomonas sp. , Pseudomonas fluorescens (19.3% of colonies) are the most numerous . Lactococcus lactis , Acetobacter indonesiensis and Moraxella osloensis bacteria also significantly contaminate dairy and fat-and-oil products . Mayonnaise is contaminated with yeast of the Pichia genus. The analysis revealed opportunistic pathogenic species: Staphylococcus warneri , Staphylococcus epidermidis, Klebsiella pneumonia , Bacillus cereus, Vibrio sp . The presented method for detecting microbial contamination using an Ion torrent PGM platform seems promising for the rapid testing of the produced dairy and fat-and-oil products.Ключевые слова
Bacteria, eukaryotic microorganisms, food products, seeding, spoilage, DNA barcoding, DNA metabarcodingВВЕДЕНИЕ
The manufacturers of dairy products, mayonnaise and other food ingredients are currently faced with the problem of contaminating products with prokaryotic and eukaryotic microorganisms that cause product damage, as well as pathogenic microorganisms. Water, industrial equipment, workers, inward raw materials, etc. may be the source for microorganisms to get into products [1]. All groups of the population are exposed to the risk of contamination by pathogenic microorganisms due to the consumption of contaminated food products, however, infants, elderly people and people who have a weakened immune system tend to the most severe consequences [2]. The list of foodborne diseases is getting constantly updated, until the 1960s, the most common food pathogens causing disease were Salmonella spp., Shigella spp., Escherichia coli, Clostridium botulinum and Staphylococcus aureus. In the 1980s and 1990s, new pathogenic species were added to this list, such as Campylobacter spp., Yersinia spp., Listeria monocytogenes, Vibrio cholera, Enterococcus faecalis and also the enterohemorrhagic strain O157 : H7 Escherichia coli [3]. The infections caused by these bacterial pathogens are now endemic in a lot of countries and cause a wide range of diseases [4]. Foodborne diseases are the result of taking the food products contaminated with pathogenic microorganisms and/or their toxins [5]. New trends in nutrition that consist in the consumption of raw and fresh food products, dry food products that have not been processed, and exotic ingredients cause a significant increase in foodborne diseases. The globalization of the food market additionally affects the outbreaks of foodborne diseases, which makes food safety a universal problem [6].
In Russia, according to Rospotrebnadzor, among other types of poisoning 147301 cases of acute intestinal infections and toxic infections caused by the defined pathogens were registered in 2016; still in 438019 cases it was not possible to define causative agents. The problem of contamination of food products by pathogenic and opportunistic microorganisms is relevant not only in the Russian Federation, but also in the world. Thus, in the USA 76000000 cases of food toxic infections have been registered for the last two decades, from which 5000 have ended with a lethal outcome. The data collection and processing system – FoodNet in the USA collects data for nine types of foodborne infections from nine states [7]. The number of confirmed cases for nine different diseases in 2002 was: salmonellosis – 6028, campylobacteriosis – 5006, shigellosis – 3875, escherichiosis – 647, cryptosporidiosis – 541, yersiniosis – 166, vibriosis – 103, listeriosis – 101, cyclosporidiosis – 43. In 1996, 11,826 cases of food poisoning caused by the E. coli 0157 : H7 bacteria were detected worldwide [8].
Milk and dairy products are among the most significant sources of foodborne diseases. For example, within the period from 1973 to 1992, 46 outbreaks of gastroenteritis caused by the consumption of raw milk were reported in the United States. Of 1733 cases, 57% were caused by Campylobacter spp., 26% – by Salmonella spp. and 2% – by E. coli 0157:H7 [9]. Moreover, L. monocytogenes [10] and Yersinia enterocolitica [11] were identified in milk. One of the most powerful outbreaks of food gastroenteritis ever recorded occurred in the Kanzai district in Japan. The source of the disease of 13420 victims was a batch of skimmed milk powder infected with the Staphylococcus aureus strain producing staphylococcal enterotoxin [12].
Of 219 samples of raw milk tested in Brazil, 16.9% contained Listeria spp. and 32.4% contained Yersinia enterocolitica [13]. The spoilage of dairy products is caused by Streptococcus spp. and Bacillus spp., which can survive even after ultra-pasteurization [14]. In cheeses the diphtheria bacteria of the Brachybacterium genus [15], as well as L. monocytogenes [16] were identified. C. tyrobutyricum is well known as the cause of amylic fermentation of cheeses [17].
The spoilage of food products high in fat, such as mayonnaise, can be caused by Pseudomonas putrefaciens [18]. Bacillus vulgatus was found in spoiled sauces [19]. When studying 17 samples of the spoiled mayonnaise, mayonnaise-like sauces and sauces with blue cheese Kurtzman et al. [20] a large amount of yeast was found in most specimens. Two thirds of the spoiled samples were seeded with Zygosaccharomyces bailii.
The standard practice of detecting bacterial pathogens is based on the cultivation of microorganisms, their microscopic observation and a biochemical analysis [21]. Although seeding with the subsequent phylogenetic analysis is the gold standard for estimating pathogenic microorganisms, the whole procedure has several limits, such as labor intensity, the duration of the analysis (5–7 days) and the complexity of the quantitative analysis. In addition, these methods are not able to detect several pathogens at the same time, so there is a need for the rapid identification of the bacterial pathogens passed through food products using high sensitivity methods [22].
A lot of authors have recently developed some methods of molecular-genetic identification based on real-time PCR using Taqman probes to identify the key pathogenic bacteria capable of seeding food products: Salmonella spp., L. monocytogenes, E. coli, thermotolerant Campylobacter spp., Yersinia enterocolitica, pathogenic Vibrio spp., Staphylococcus aureus and pathogenic Clostridia spp. [23].
DNA barcoding has recently become increasingly popular. DNA barcoding is used as an instrument of taxonomic identification of organisms [24–26]. This approach consists in sequencing the gene site and comparing the obtained sequence with those that are already available in international genetic databases, such as Boldsystem and GenBank. The gene of subunit 1 of cytochrome oxidase of mitochondrial DNA for animals [27], the gene of the internal transcriptional spacer (ITS) of nuclear DNA for fungi [28] and the genes of rbcLb and matK for plants are most often used as such a gene [29]. The limit of this method is complexity in the analysis of a substrate that has the mixtures of DNA of the organisms of different taxonomic groups. A more advanced approach is the so-called DNA metabarcoding, which is performed using next generation sequencers (NGS sequencing). In this case, the analysis of complex biological mixtures is possible. The next-generation sequencing revolutionized food microbiology by developing new high-performance technologies, such as 16S rRNA microbiological profiling and shotgun sequencing, which were used to study the microbiota composition of various food products [30].
At present, the manufacturers of dairy and fat-andoil products of Russia are obliged to comply with the requirements of the Customs Union for food safety, in particular, the products should not contain pathogenic microorganisms. There is also a limitation of the number of yeast and mold microorganisms and the total number of bacteria. The purpose of this study was to analyze the ready-to-eat commercial products of the dairy and fat-and-oil industry produced in Russia, for their microbiological composition, using classical DNA barcoding, as well as DNA metabarcoding.
ОБЪЕКТЫ И МЕТОДЫ ИССЛЕДОВАНИЯ
Study objects. As the study object, the food dairy and fat-and-oil products produced in Russia were used (Table 1). 1 g of a solid or 1 ml of a liquid solution was taken for the study.
Microbiological inoculation. To check the
bacterial content of food products for different
physiological groups of microorganisms, seeding was
carried out in the nutrient media of the following
composition:
(1) FPA (fish-peptone agar) (to determine the total
microbial number of mesophilic, aerobic and facultative
anaerobic microorganisms): pancreatic fishmeal
hydrolyzate – 12 g/l; enzymatic peptone – 12 g/l;
NaCl – 6 g/l; microbiological agar – 10 g/l; pH 7.1–7.5.
(2) Giss-GRM medium (to identify enterobacteria):
pancreatic fishmeal hydrolyzate – 6 g/l; NaCl – 3.5 g/l;
Na2HPO4 – 0.2 g/l; mannitol – 3.5 g/l; blue thymol
bromine – 0.04 g/l; microbiological agar – 3,5 g/l; pH 7.4.
(3) GRM nutrient medium No.2 (Saburo) (to detect
yeast and mold): pancreatic fishmeal hydrolyzate –
10 g/l; pancreatic casein hydrolyzate – 10 g/l; yeast
extract – 2 g/l; NaH2PO4 – 2 g/l; D-glucose – 40 g/l;
microbiological agar – 10 g/l; pH 6.0. To suppress the
growth of the extraneous microflora, 10 ml of 1%
chloramphenicol solution per 1 liter of the medium
were added to the medium before seeding.
CFU (Colony forming units: the indicator of the number of viable microorganisms per volume unit) were counted using the method of 10-time limiting dilutions in three biological repetitions. In accordance with the requirements of ASTM D5465-93 (2012) (GOST 26670-91) Petri dishes with the amount of CFU from 30 to 300 were counted.
DNA barcoding. DNA was isolated from the colonies of microorganisms using the Probe-GS kit (DNA technology, Russia) according to the attached instructions. The polymerase chain reaction was carried out using Taq polymerase with a Mastercycler personal device (Eppendorf, Germany). The following components were mixed in a 0.25 ml tube: a 10X reaction buffer – 2.5 μl; 10 mM dNTP – 1 μl; a 10 μmol primer – 1 μl; a 10 μmol reverse primer – 1 μl; 25 mM Mg2+ – 3 μl; a matrix – 1 μg; thermostable Taqpolymerase – 2.5 units; deionized water – up to 25 μl. The following temperature cycles were used: 3 min at 94°C, 35 cycles, 30 sec at 94°C, 30 sec at 54°C, 45 sec at 72°C and the final elongation for 10 min at 72° C. The following were used as primers: to amplify bacterial DNA: direct 785F GGATTAGATACCCTGGTA, reverse 1492R TACGGYTACCTTGTTACGACTT; for the amplification of fungal DNA: direct ITS1 TCCGTAGGTGAACCTGCGG, reverse ITS2 GCTGCGTTCTTCATCGATGC and ITS4 TCCTCCGCTTATTGATATGC. The PCR products were visualized by means of electrophoresis in 2% agarose gel. Ethidium bromide was used as nucleic acid dye. The size of the products was determined by comparison with the DNA markers of the known length (Evrogen, Russia).
The extraction from agarose gel and amplicon purification was performed using a commercially available Cleanup Standard kit (Evrogen, Russia). The purified PCR products were sequenced using an Applied Biosystems 3500 genetic analyzer with a BigDye Terminator v3.1 Cycle Sequencing Kit.
Enrichment of microorganisms in food substrates. The pre-enrichment of microorganisms for the subsequent analysis using the method of highperformance sequencing was performed in a sterile nutrient medium of the following composition: 2% glucose and 1% peptone. 1 g (or 1 ml) of food substrate was introduced into a medium of 9 ml in volume and incubated for 24 hours at 27°C. The food substrate: enrichment medium ratio was taken in accordance with the instructions (MUK 4.2.2872-11). Then, 100 μl of the medium was isolated for DNA isolation. When DNA was being isolated from mayonnaise and mayonnaise sauces, an additional step was taken, which consisted in the precipitation of microorganism cells by means of centrifugation at 10,000 g for 5 minutes. The supernatant was removed together with the fat, and the precipitate was resuspended in 100 μl of sterile water and used to isolate DNA. This stage is necessary to remove fats that prevent the release of DNA.
High-performance sequencing. The DNA of microorganisms was isolated using a Probe-GS kit (DNA technology, Russia) according to the attached instructions. The multiplex PCR was performed using the primers 785F/1492R and ITS1/ITS2 (see above). The PCR products were purified by magnetic particles AMPure XP Beads (Beckman Coulter, USA). The purified product was used to prepare sequencing libraries using The Ion AmpliSeq Library Kit 2.0 according to the kit protocol. The Ion Xpress Barcode Adapters (Thermo Fisher Scientific, USA) was used to barcode the samples. To determine the concentration of libraries, a real-time PCR kit (The Library Quantification Kit Ion Torrent Platforms (Kapa Biosystems)) was used.
The sequencing was performed using an IonTorrent PGM platform with an Ion PGM Hi-Q View Sequencing Kit together with the system Ion OneTouch 2 and an Ion PGM Hi-Q View OT2 Kit (Thermo Fisher Scientific, USA).
The source material for the bioinformatic analysis
was bam files containing the source information about
the reads obtained during DNA sequencing. The bam
files containing nucleotide sequences in the binary form
were converted to the FASTQ format using a samtools
Version 1.2 package [31]. The demultiplexing was
performed using the application fastq-multx, which is
part of the software package ea-utils, Version 1.3 [32].
For this purpose, a file was prepared containing the
sequences of primers that are part of the amplicons of
different origin and the following command was
executed:
fastq-multx –B barcodes.fa <имя_файла. fastq> -m 5 -o
%.fastq.
The -m 5 argument resolves up to 5 discrepancies
between the primer sequence and the initial amplicon
sequence in a FASTQ file. This parameter has been
obtained empirically and does not affect the accuracy
of the further analysis, making it possible at the same
time to reduce the number of reads discarded as
unidentified. The fitting of reads was performed for
each sample using the command:
-
trunclength <длина_рида> -fastqout
<имя_файла_длина.fastq>.
Further on, the reads were filtered according to
reading quality based on the expected number of
errors [33]:
usearch -fastq_filter <имя_файла_длин. fastq> -
fastq_maxee 1.0 -fastaout <имя_файла_filtered. fasta>.
Before searching for OTU (Operational taxonomic
unit), unique sequences were identified:
usearch -fastx_uniques <имя_файла_filtered.fasta> -
fastaout <имя_файла_uniques.fasta>
-sizeout -relabel Uniq.
Two different approaches were used to search for OTU. The former is based on the UNOISE2 algorithm [34], the task of which is to reduce the noise level in the sample by correcting errors. The result of its application is the isolation of all biologically correct sequences in a set of reads: usearch -unoise3 <имя_файла_uniques.fasta> -zotus <имя_файла_ zotus.fasta>.
The read filtering, searching for unique sequences and clustering to obtain OTU were performed using a USEARCH software package version 10.0.240 [35]. The species of microorganisms contained in the sample was identified using a SILVA database (https://www.arbsilva.de/) in the case of bacterial DNA, and a BOLD Systems database (http://www.boldsystems.org/ index.php/IDS_OpenIdEngine) in the case of DNA of eukaryotic microorganisms.
РЕЗУЛЬТАТЫ И ИХ ОБСУЖДЕНИЕ
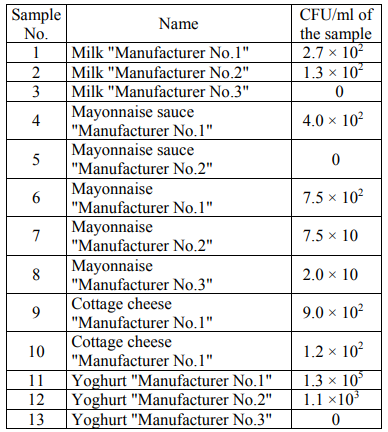
In the course of microbiological analysis, the amount of the colonies grown in nutrient media was initially estimated. Tables 2 present the number of the colonies grown in an FPA nutrient medium (CFU/ml) for the studied products.
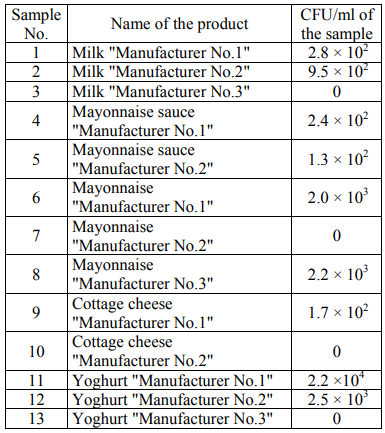
It was impossible to calculate the number of colonies due to their high concentration in the sample of yoghurt "Manufacturer No.1" in its initial dilution. Table 3 presents the number of the colonies grown in a Giss nutrient medium (CFU/ml) for the studied products.
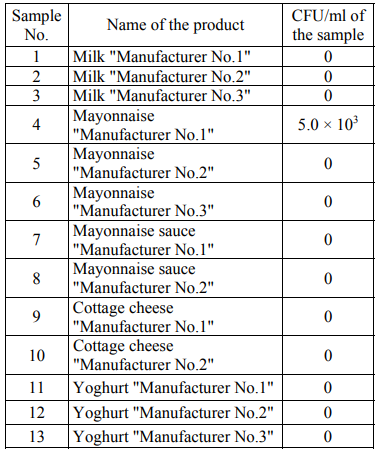
In general, the number of the colonies of microorganisms grown in the Giss medium was less than that in the FPA medium. To determine the number of eukaryotic microorganisms, the Saburo medium was used with the addition of an antibiotic (see materials and methods). Tables 4 present the number of the colonies grown in the Saburo nutrient medium (CFU/ml) for the studied products.
Filamentous fungi were not detected in any of the samples. Contaminating with yeast was found in mayonnaise "Manufacturer No.1".
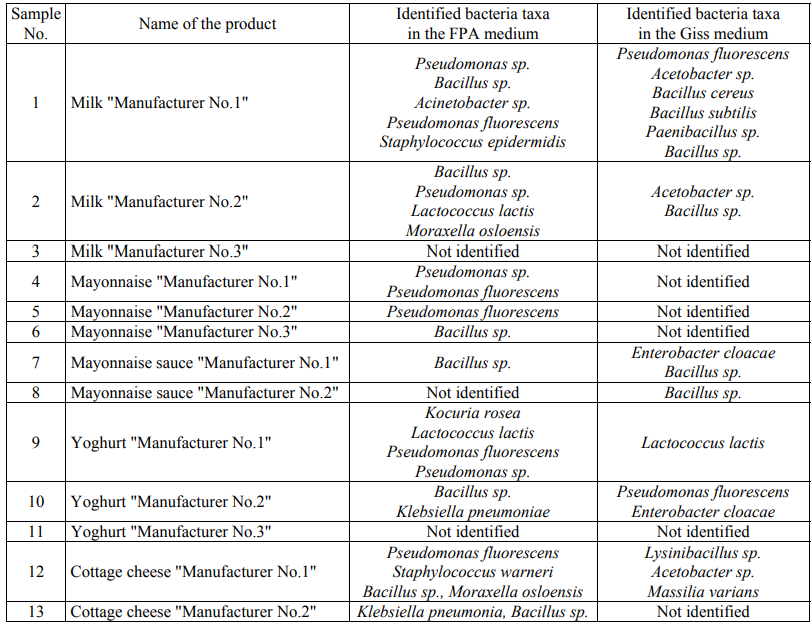
In aseptic conditions, the grown colonies were selected, DNA was isolated from them and the amplification of the 16S rRNA gene in bacteria or the amplification of the DNA segment involving the genes 18S rRNA, 5.8S rRNA and 28S rRNA and the intergenic segments ITS1 and ITS2 in eukaryotic microorganisms were carried out. The amplicons were then isolated and sequenced using the Sanger method. A total of 117 nucleotide sequences were obtained. The obtained nucleotide sequences were compared with those already available in the international GenBank database. Table 5 presents the identified taxa of microorganisms.
The data on the number of microorganisms in the FPA medium are often in agreement with the data obtained in the Giss medium. In some samples, a difference in the number of bacteria in the FPA medium and in the Giss medium was revealed. Thus, in the FPA medium, when seeding the samples of mayonnaise sauce "Manufacturer No.2" and yoghurt "Manufacturer No.3" CFU is 0, and in the Giss medium with mannitol CFU/ml is from 102 to 103. In the Giss medium, when seeding the samples of mayonnaise "Manufacturer No.2", yoghurt "Manufacturer No.1" and cottage cheese "Manufacturer No.2" CFU/ml is 0, and in the FPA medium CFU/ml is from 105 to 106 for a number of samples.
It should be noted that the representatives of the genera Pseudomonas, Bacillus and Lactococcus have been identified in both media. The seeding in the FPA nutrient media and the Giss medium made it possible to identify the differences at the genus level: the seeding in the FPA medium made it possible to identify the representatives of the genera Kocuria, Staphylococcus, Moraxella, Paucisalibacillus, Acinetobacter and Klebsiella, while the representatives of the genera Paenibacillus, Lysinibacillus, Enterobacter, Acetobacter and Massilia were identified in the Giss medium.
Filamentous fungi and yeast were not detected in any of the samples except for the mayonnaise sample of Manufacturer No.1. According to the sequence of the DNA segment that includes the genes 18S rRNA, 5.8S rRNA and 28S rRNA and the intergenic segments ITS1 and ITS2, this yeast has been identified as the representatives of Saccharomycetes of the Saccharomycetaceae family of the Pichia genus.
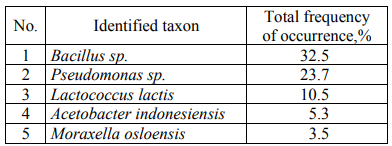
Table 6 presents the quantitative ratios of the identified microorganisms.
When analyzing the quantitative ratio of microorganisms, it was revealed that Bacillus sp., among which it is possible to distinguish conditionally Bacillus licheniformis (16.67% of colonies) (the taxon closest to DNA in the international GenBank database) and Bacillus subtilis (11.4% of colonies) most often contaminate dairy and fat-and-oil products. Among Pseudomonas sp., Pseudomonas fluorescens (19.3% of colonies) are the most numerous.
To study the method of high-performance sequencing in order to identify microbiological contamination of fat-and-oil products, we analyzed 3 samples of milk, 3 samples of mayonnaise, 2 samples of mayonnaise sauce and 2 samples of cottage cheese. After the preliminary enrichment of the studied samples of food products, 100 μl of broth was selected (or the precipitate was resuspended in 100 μl of broth, if the isolation was made from mayonnaise or mayonnaise sauce). Then DNA was isolated and PCR was performed. Since one sample that corresponds to one barcode of the sequencer, contained several types of the amplicons obtained from multiplex PCR (the fragments of bacterial 16S rRNA and a segment that includes the genes 18S rRNA, 5.8S rRNA and 28S rRNA, and also the intergenic segment ITS1 in fungi), the demultiplexing of reads was carried out before the further analysis (see materials and methods). As a result, two new reads that contain the DNA sequences of bacteria and fungi were obtained, respectively, for each source file.
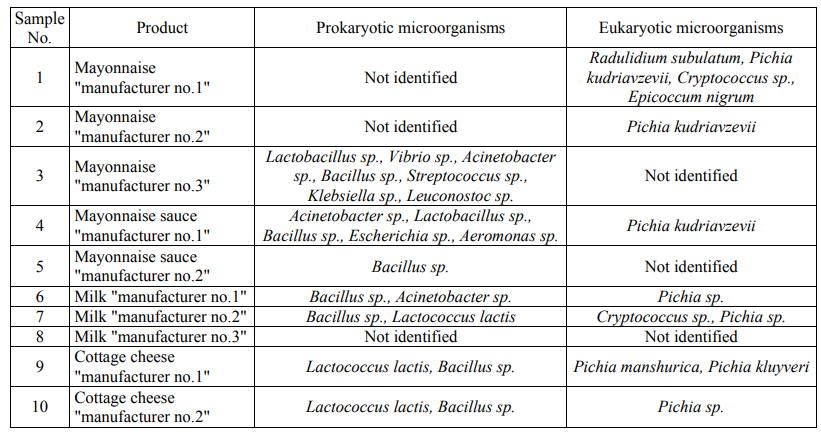
Table 7 presents the sequence coincidences obtained during high-performance sequencing with the sequences of prokaryotic and eukaryotic microorganisms available in the international databases.
When analyzing the taxa revealed by means of highperformance sequencing, it turned out that the list of microorganisms in dairy products is less extensive than in the case of identification by classical DNA barcoding. While there was a different situation .... high fat products (mayonnaise and mayonnaise sauces). This effect is most likely due to the additional concentration of microorganisms when enriching mayonnaise and mayonnaise sauces, since when studying these products using classical DNA barcoding there was no concentration and the products were immediately introduced into nutrient media. Besides, the additional optimization is probably required when preparing DNA libraries from dairy products, as well as, possibly, the targeted amplification of target microorganism groups. The high-performance sequencing has also better revealed the presence of eukaryotic microorganisms in the studied food products.
The analysis of species composition of microorganisms using classical DNA barcoding, as well as metabarcoding, allowed us to identify opportunistic pathogenic species from the number of products: Staphylococcus warneri, Staphylococcus epidermidis, Klebsiella pneumonia, Bacillus cereus, Vibrio sp. Klebsiella pneumoniae is one of the causative agents of pneumonia, as well as some urogenital infectious diseases and purulent abscesses of the spleen and liver. This microorganism can also be pathogenic to some animals. Some strains are multiresistant to antibiotics, the capsule is a virulence factor [36–38].
Staphylococcus warneri, like other staphylococci, belongs to the 4th group of pathogenicity, it rarely causes human and animal diseases, it is mainly characteristic of the patients with a blunt immunity. There are cases of conjunctivitis, the infections of the urogenital tract and septicemia associated with S. warneri [39, 40].
Staphylococcus epidermidis occurs on the mucous membranes and human skin, can cause endocarditis, sepsis, the purulent infection of wounds and urinary tract infections [39].
Bacillus cereus is a dangerous pathogen that causes foodborne toxic infections in human (including diarrhea and the emetic syndrome). The diarrheal syndrome is caused by a high molecular weight peptide toxin, whereas the emetic syndrome is caused by a low molecular weight thermostable toxin [36, 41].
Vibrio sp. identified by high-performance sequencing, also poses a risk to human health, at the same time, some species of this genus are even related to the 2nd and 3rd group of pathogenicity [41, 43].
In addition to pathogenic microorganisms, the microorganisms that spoil products were detected. The microbiological spoilage of products – the development of harmful microorganisms in food products with the subsequent accumulation of their waste products – can also be dangerous because of the evolved toxins and the development of pathogenic microbiota. The key cause of microbiological spoilage of products is fermentation. Most often, the spoilage of products is caused by acetic and amylic fermentation, as well as rotting [44]. Acetic fermentation forms the Acetobacter bacteria genus, which we detected in milk and cottage cheese. These bacteria are able to oxidize ethanol to acetic acid, acetate and lactate. Acetobacter is of particular importance for the food industry, since these bacteria spoil products, producing acetic acid or ethyl acetate. The substrates that contain sugar or fruit are particularly susceptible to acetic acid fermentation [45].
In the course of the analyzes made, we found the presence a large amount of bacteria of the Bacillus genus the reproduction of which can lead to the decay of food substrates. The rotting process is caused by putrefactive microorganisms that are widespread in all habitats, including animals and plant organisms. The deepest breakdown of proteins is caused by the representatives of the Enterobacteriaceae family (for example, the genera Proteus and Escherichia) and the spore-forming bacteria of the genus Bacillus and Clostridium [46].
With the help of high-performance sequencing, we managed to identify eukaritic microorganisms in several samples, while with the help of microbiological seeding, eukaryotic microorganisms were only detected in one of the samples. This can be explained by the higher sensitivity of new generation sequencing when identifying microorganisms compared to microbiological seeding or dead eukaryotic microorganisms were possibly identified, since their DNA can be retained for a long time in food substrates. In our study we used the method of classical DNA barcoding (the sequencing of marker genes of prokaryotic and eukaryotic microorganisms), as well as the method of DNA metabarcoding based on highperformance sequencing on an Ion torrent PGM platform. The main advantage of the method of classical DNA barcoding is the lower cost of analysis in relation to metabarcoding. The main disadvantage of the method of classical DNA barcoding is the requirement for the homogeneity of biological material, which is achieved either by the preliminary separation of the studied microorganisms or by the molecular cloning of PCR fragments. In our case this requirement was achieved by means of the preliminary seeding of microorganisms in solid nutrient media. When performing metabarcoding based on high-performance sequencing, the preliminary seeding is not required.
The next-generation sequencing (NGS) collectively describes several technologies that provide the mass parallel sequencing of heterogeneous DNA fragments. With regard to the monitoring of the microbial community, these fragments consist of the short segments amplified using the universal primers targeted at the known marker genes, predominantly the prokaryotic 16S rRNA and fungal ITS genes. At present, two NGS systems are basically used for profiling microbial communities; these are the sequencing platforms 454 Life Sciences pyrosequencing [47] and Illumina [48]. We have shown that the Ion torrent PGM platform is also capable to identify effectively the complex mixtures of microorganisms. The presented detection method using an Ion torrent PGM platform seems promising for rapid microbial characteristics in multiple food samples with the unknown composition of microorganisms by determining numerous nucleic sequences simultaneously without the need for cloning PCR products and microbiological seeding. However, it should be noted that high-performance sequencing is an expensive and time-consuming method yet [49].
The microorganisms that we have identified, which belong to the pathogenic group in ready-made dairy and fat-and-oil products, are a wakeup call. The production control at the enterprises that produced the studied products was probably carried out using only the microorganisms of the 3rd group of pathogenicity, such as Shigella spp. and Listeria monocytogenes. However, the microorganisms of the 4th pathogenicity group (in particular, Bacillus cereus, Staphylococcus sp., Klebsiella pneumoniae and Vibrio sp.) that we have detected, are not equally allowed in ready-made dairy and fat-and-oil products, as, for example, Shigella spp. and Listeria monocytogenes (TR TS 033/2013 "On safety of milk and dairy products" and TR TS 024/2011 "Technical Regulations on the safety of fat-and-oil products"), since they also belong to pathogenic microorganisms, although belong to the last 4 group of the list of pathogenic microorganisms (SP 1.3.2322-08 "Safety of work with microorganisms"). The technical regulations of the customs union do not allow the presence of pathogenic microorganisms in any food products, the presence of coliforms is also not allowed or is sharply limited, the limitations of the amount of yeast and mold are introduced. Thus, for example, TR TS 033/2013 "On safety of milk and dairy products" separately prescribes the non-admissibility of the presence of the coliforms S.aureus, L. monocytogenes and Salmonella spp., while for a number of dairy products the counting of eukaryotic microorganisms is not regulated, but for other products, the amount of yeast and mold should not exceed 50 per 1 cm3 (g). In the international documents that regulate the microbiological standards of food products, for example, European Hygiene and Food Safety Criteria 2073/2005, there is also a ban on the content of pathogenic microorganisms, especially L. monocytogenes, Cronobacter spp. (Enterobacter sakazakii) and Salmonella spp.
In general, the methods that we have presented on the basis of DNA metabarcoding can become an alternative to standard microbiological seeding during production control at the food enterprises of the dairy and fat-and-oil industry, since, despite the high cost of high-performance sequencing, the analysis time is significantly reduced, which will allow to faster load up store shelves with the products with a short shelf life.
БЛАГОДАРНОСТИ
The study has been supported by the Ministry of Education and Science of the Russian Federation (Agreement No.14.577.21.0257, the unique agreement code is RFMEFI57717X0257)
СПИСОК ЛИТЕРАТУРЫ
- Jay J.M., Loessner M.J., and Golden D.A. Modern Food Microbiology, 7th edn. New York: Springer, 2005. 782 p.
- The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA Journal, 2015, vol. 13, no. 1, pp. 1-165. DOI: 10.2903/j.efsa.2015.3991.
- Newell D.G., Koopmans M., Verhoef L., et al. Food-borne diseases - The challenges of 20years ago still persist while new ones continue to emerge. International Journal of Food Microbiology, 2010, vol. 139, no. 1, pp. 3-15. DOI: 10.1016/j.ijfoodmicro.2010.01.021.
- Ranjbar R., Rahbar M, Naghoni A., et al. A cholera outbreak associated with drinking contaminated well water. Archives Iranian Medicine, 2011, vol. 14, no. 5, pp. 339-340.
- Ferrario C., Lugli G.A., Ossiprandi M.C., et al. Next generation sequencing-based multigene panel for high throughput detection of food-borne pathogens. International Journal of Food Microbiology, 2017, vol. 256, pp. 20-29. DOI: 10.1016/j.ijfoodmicro.2017.05.001.
- Scallan E., Hoekstra R.M., Mahon B.E., Jones T.F., and Griffin P.M. An assessment of the human health impact of seven leading foodborne pathogens in the United States using disability adjusted life years. Epidemiology and Infection, 2015, vol. 143, no. 13, pp. 2795-2804. DOI: 10.1017/S0950268814003185.
- Centers for Disease Control and Prevention. Outbreaks of Salmonella serotype Enteritidis infection associated with eating raw or undercooked shell eggs - United States, 1996-1998. Morbidity And Mortality Weekly Report, 2000, vol. 49, no. 4, pp. 73-79.
- Machino H., Araki K., Minami S., et al. Recent outbreaks of infections caused by Esherichia coli O157:H7 in Japan. In Escherichia coli O157:N7 and Other Shiga Toxin-Producing E. coli Strains. Washington DC: ASM Press. 1998, pp. 73-81.
- Headric M.L., Korangy S., Bean N.H., et al. The epidemiology of raw milk-associated foodborne disease outbreaks reported in the United States, 1973 through 1992. American Journal of Public Health, 1998, vol. 88, no. 8, pp. 1219-1221. DOI: 10.2105/AJPH.88.8.1219.
- Gitter M., Bradley R., and Blampied P.H. Listeria monocytogenes infection in bovine mastitis. Veterinary Record, 1980, vol. 107, no. 17, pp. 390-393.
- Moustafa M.K., Ahmed A.A-H., Marth E.H. Occurrence of Yersinia enterocolitica in raw and pasteurized milk. Journal of Food Protection, 1983, vol. 46, no. 4, pp. 276-278. DOI: 10.4315/0362-028X-46.4.276.
- Asao T., Kumeda Y., Kawai T., et al. An extensive outbreak of staphylococcal food poisoning due to low-fat milk in Japan: Estimation of enterotoxin A in the incriminated milk and powdered skim milk. Epidemiology and Infection, 2003, vol. 130, no. 1, pp. 33-40. DOI: 10.1017/S0950268802007951.
- Tibana A., Warnken M.B., Nunes M.P., Ricciardi I.D., and Noleto A.L.S. Occurrence of Yersinia species in raw and pasteurized milk in Rio de Janeiro, Brazil. Journal of Food Protection, 1987, vol. 250, no. 7, pp. 580-583. DOI: 10.4315/0362-028X-50.7.580
- Pettersson B., Lembke F., Hammer P., Stackebrandt E., and Priest E.G. Bacillus sporothermodurans, a new species producing highly heat-resistant endospores. International Journal of Systematic Bacteriology, 1996, vol. 46, pp. 759-764. DOI: 10.1099/00207713-46-3-759.
- Schubert K., Ludwig W., Springer N., et al. Two coryneform bacteria isolated from the surface of French Gruyure and Beaufort cheeses are new species of the genus Brachybacterium: Brachybacterium alimentarium sp. nov. and Brachybacterium tyrofermentans sp. nov. International Journal of Systematic Microbiology, 1996, vol. 46, no. 1, pp. 81-87. DOI: 10.1099/00207713-46-1-81.
- Rudolf M. and Scherer S. High incidence of Listeria monocytogenes in European red smear cheese. International Journal of Food Microbiology, 2001, vol. 63, nos 1-2, pp. 91-98. DOI: 10.1016/S0168-1605(00)00413-X.
- Klijn N., Nieuwenhof F.F.J., Hoolwerf J.D., Van der Waals C.B., and Weerkamp A.H. Identification of Clostridium tyrobutyricum as the causative agent of late blowing in cheese by species PCR amplification. Appled and Environmental Microbiology, 1995, vol. 61, no. 8, pp. 2919-2924.
- Hugo C.J., Segers P., Hoste B., Vancanneyt M., and Kersters K. Chryseobacterium joostei sp. nov., isolated from the dairy environment. International Journal of Systematic and Evolutionary Microbiology, 2003, vol. 53, pp. 771-777. DOI: 10.1099/ijs.0.02232-0.
- Pederson C.S. Bacterial spoilage of a thousand island dressing. Journal of Bacteriology, 1930, vol. 20, no. 2, pp. 99-106.
- Kurtzman C.P., Rogers R., and Hesseltine C.W. Microbiological spoilage of mayonnaise and salad dressings. Applied Microbiology, 1971, vol. 21, no. 5, pp. 870-874.
- Mao Z., Zheng H., Wang X., et al. DNA microarray for direct identification of bacterial pathogens in human stool samples. Digestion, 2008, vol. 78, no. 2-3, pp. 131-138. DOI: 10.1159/000174465.
- Ranjbar R., Karami A., Farshad S., Giammanco G.M., and Mammina C. Typing methods used in the molecular epidemiology of microbial pathogens: a how-to guide. New Microbiologica, 2014, vol. 37, no. 1, pp. 1-15.
- Rodriguez-Lazaro D. (ed.). Real-Time PCR in Food Science: Current Technology and Applications. Norfolk, UK: Caister Academic Press., 2013. 285p.
- Hebert P.D. and Gregory T.R. The promise of DNA barcoding for taxonomy. Systematic Biology, 2005, vol. 54, no. 5, pp. 852-859. DOI:10.1080/10635150500354886.
- Hebert P.D., Cywinska A., Ball S.L., and DeWaard J.R. Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences, 2003, vol. 270, no. 1512, pp. 313-321. DOI: 10.1098/rspb.2002.2218.
- Ferri G., Alù M., Corradini B., Licata M., and Beduschi G. Species identification through DNA "barcodes". Genetic testing and molecular biomarkers, 2009, vol. 13, no. 3, pp. 421-426. DOI: 10.1089/gtmb.2008.0144.
- Hebert P.D., Ratnasingham S., and DeWaard J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal B: Biological Sciences, 2003, vol. 270, no. 1, pp. 96-99.
- Schoch C.L., Seifert K.A., Huhndorf S., et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America, 2012, vol. 109, no. 16, pp. 6241-6246. DOI: 10.1073/pnas.1117018109.
- Dong W., Cheng T., Li C., et al. Discriminating plants using the DNA barcode rbcLb: An appraisal based on a large data set. Molecular Ecology Resources, 2014, vol. 14, no. 2, pp. 336-343. DOI: 10.1111/1755-0998.12185.
- Mayo B., Rachid C.T., Alegría A., et al. Impact of next generation sequencing techniques in food microbiology. Current Genomics, 2014, vol. 15, no. 4, pp. 293-309. DOI: 10.2174/1389202915666140616233211.
- Li H., Handsaker B., Wysoker A., et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics, 2009, vol. 25, no. 16, pp. 2078-2079. DOI: 10.1093/bioinformatics/btp352.
- Aronesty E. Comparison of sequencing utility programs. Open Bioinformatics Journal, 2013, vol. 7, no. 1, pp. 1-8. DOI: 10.2174/1875036201307010001.
- Edgar R.C. and Flyvbjerg H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics, 2015, vol. 31, no 21, pp. 3476-3482. DOI:10.1093/bioinformatics/btv401.
- Edgar R.C. UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv, 2016. DOI: https://doi.org/10.1101/081257.
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 2010, vol. 26, no. 19, pp. 2460-2461. DOI: 10.1093/bioinformatics/btq461.
- Korotyaev A.I. and Babichev S.A. Meditsinskaya mikrobiologiya, immunologiya i virusologiya [Medical microbiology, immunology and virology]. St. Petersburg: SpetsLit Publ., 2008. 767 p.
- Pozdeev O.K. Meditsinskaya mikrobiologiya [Medical microbiology]. Moscow: Geotar-Media Publ., 2001. 778 p.
- Ryabkova E.L. Optimizatsiya antibiotikoterapii nozokomial'nykh infektsiy, vyzvannykh Klebsiella pneumonia, v statsionarakh Rossii [Optimization of antibiotic therapy of nosocomial infections caused by Klebsiella pneumonia in hospitals in Russia]. Smolensk: n. publ., 2006. 23 p.
- Akatov A.K. and Zueva V.S. Stafilokokki [Staphylococci]. Moscow: Meditsina Publ., 1983. 242 p.
- Kamath U., Singer C., and Isenberg H.D. Clinical significance of Staphylococcus warneri bacteremia. Journal Clinical Microbiology, 1992, vol. 30, no. 2, pp. 261-264.
- Vasilyev D.A., et al. Identifikatsiya bakteriy Bacillus cereus na osnove ikh fenotipicheskoy kharakteristiki [Identification of Bacillus cereus bacteria based on their phenotypic characteristic]. Ulyanovsk: SRICMB of Ulyanovsk SAA Publ, 2013. 98 p.
- Newton A., Kendall M., Vugia D.J., Henao O.L., and Mahon B.E. Increasing rates of vibriosis in the United States, 1996-2010: review of surveillance data from 2 systems. Clinical Infectious Diseases, 2012, vol. 54, pp. 391-395. DOI: 10.1093/cid/cis243.
- Mead P.S., Slusker L., Dietz V., et al. Food-related illness and death in the United States. Emerging Infectious Diseases, 1999, vol. 5, pp. 607-625.
- Gustavsson J., Cederberg C., Sonesson U., Otterdijk R., and Meybeck A. Global food losses and food waste - Extent, causes and prevention. Rome: FAO, 2011. 38 p.
- Tesfaye W., Morales M.L., Garcıa-Parrilla M.C., and Troncoso A.M. Wine vinegar: Technology, authenticity and quality evaluation. Trends Food Science and Technology, 2002, vol. 13, no. 1, pp. 12-21. DOI: 10.1016/S0924-2244(02)00023-7.
- Mitsuoka T. Development of functional foods. Bioscience of Microbiota, Food and Healths, 2014, vol. 33, no. 3, pp. 117-128. DOI: 10.12938/bmfh.33.117.
- Margulies M., Egholm M., Altman W.E., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature, 2005, vol. 437, no 7057, pp. 376-380. DOI: 10.1038/nature03959.
- Balasubramanian S. Solexa sequencing: Decoding genomes on a population scale. Clinical Chemistry, 2015, vol. 61, no. 1, pp. 21-24. DOI: 10.1373/clinchem.2014.221747.
- Humblot C. and Guyot J.P. Pyrosequencing of tagged 16S rRNA gene amplicons for rapid deciphering of the microbiomes of fermented foods such as pearl millet slurries. Applied and Environmental Microbiology, 2009, vol. 75, no. 13, pp. 4354-4361. DOI: 10.1128/AEM.00451-09.