Аннотация
Wines DNA authentication is a technological process of their authenticity verification by genetic identification of the main plant ingredient by means of molecular genetic analysis of the residual amounts of Vitis vinifera L nucleic acids extracted from end product cellular debris. The main aim of the research was the analysis of scientific and methodological approaches to the extraction of residual amounts of nucleic acids in wine raw materials and DNA authentication of wines for their subsequent application in solving the problem of determining wine products authenticity and place of origin. The prior art includes various approaches to the extraction of Vitis vinifera L. nucleic acids among which the three methods by Savazzini & Martinelli, Pereira and Bigliazzi can be named basically. Analysis of the effectiveness of different methods of DNA extraction from wines indicates the superiority of the Pereira method over other traditional methods of extraction in terms of DNA yield and quality. Besides, the nucleic acid extracted from wines is characterized as residual since its concentration is significantly reduced in a multi-stage wine production process. The yield of extracted nucleic acid also decreases as the wine ages. The use of microsatellite DNA loci designed for grapes genetic identification is one of the approaches applicable for wine DNA authentication.Ключевые слова
Wine, grapes, variety, Vitis vinifera L, DNA, authentication, identification, marker, SSR, SNP, PCR, HRM analysisВВЕДЕНИЕ
Considering its wide assortment variety and multicomponent chemical composition, wine production belongs to the segment of difficult-to- identify goods whose established authenticity serves as a basic element of consumers and producers’ rights protection [1, 2].
The search for objective identification criteria with a high degree of wine products authenticity and place of origin assessment reliability is a strategically important task achievable by multidisciplinary science- intensive approaches [3, 4].
The complex identification scheme based on the approved methods of analysis (documentary, visual, organoleptic, and physicochemical) [5–9] can be expanded by the molecular genetic method [10] applicable for wine DNA authentication [11, 12].
Wine DNA authentication is a technological process of authenticity verification by genetic identification of the main plant ingredient – wine grapes – by means of molecular genetic analysis of the residual amounts of Vitis vinifera L nucleic acids extracted from end product cellular debris [11, 12].
Analysis of research and methodological approaches to the extraction of the residual amounts of nucleic acids in wine raw materials, and wine DNA authentication demonstrates the applicability of DNA technologies for the monitoring of counterfeit and adulterated wine products.РЕЗУЛЬТАТЫ И ИХ ОБСУЖДЕНИЕ
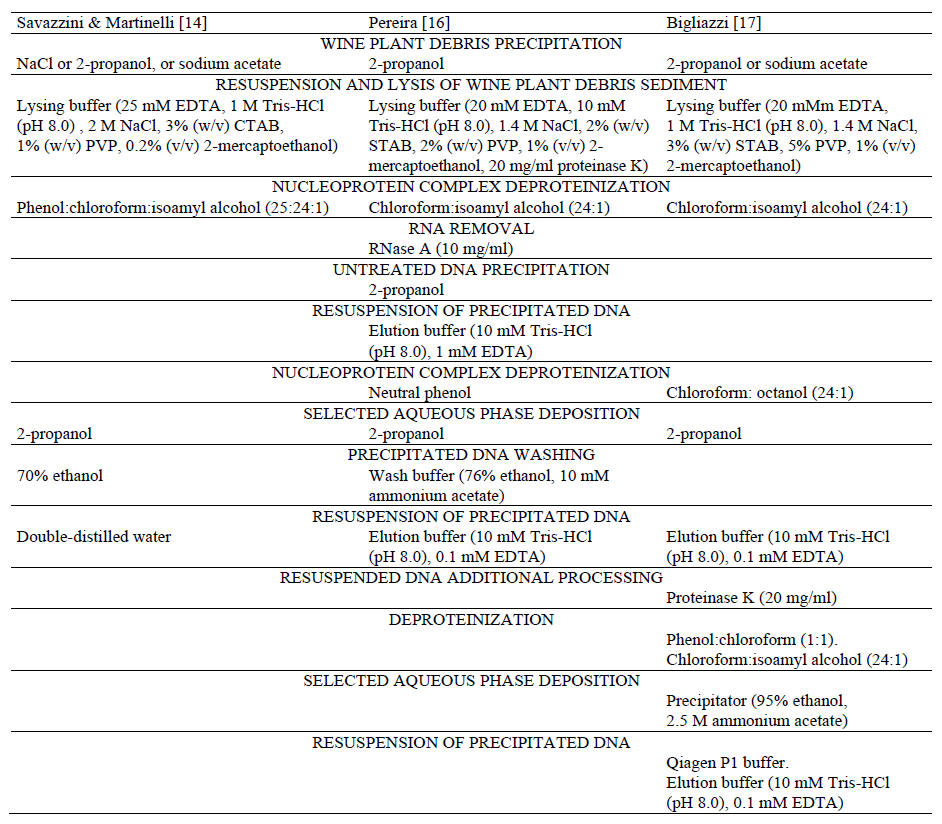
Methods of extraction of wine raw materials DNA residual amounts. The prior art includes various approaches to the extraction of Vitis vinifera L. nucleic acids [13–21], among which the three methods by Savazzini & Martinelli [14], Pereira [16] and Bigliazzi [17] can be named basically in both the original and modified set-ups [11].
Reagents used at various stages of sample preparation and extraction of Vitis vinifera L. DNA in the three namesake methods of nucleic acids extraction from wines are shown in Table 1.
The unifying feature of the methods presented is the precipitation of wine plant debris by centrifugation using precipitators, suchas sodium chloride, 2-propanol, and sodium acetate separately or in combination of the latter two.
Resuspension of the plant debris sediment and its lysis are carried out by a multicomponent lysing buffer consisting of ethylenediaminotetraacetic asid (EDTA), trisaminomethane hydrochloric acid (Tris-hcl), sodium chloride (NaCl), cetyltrimethylammonium bromide (CTAB), polyvinylpyrrolidone (PVP), and 2-mercaptoethanol [14, 16, 17]. The buffer also includes the proteinase K in the Pereira method [16].
The stage of deproteinization of the lysate nucleoprotein complex is carried out by organic solvents (chloroform and isoamyl alcohol), with the inclusion of phenol in the three-component mixture phenol:chloroform:isoamyl alcohol at the ratio 25:24:1 in the Savazzini & Martinelli method [14].
The Pereira method [16] includes the stage of RNA removal by treatment with RNase A followed by untreated DNA deposition by centrifugation, further resuspension of the deposited DNA with an elution buffer and additional deproteinization with neutral phenol. In the Bigliazzi method [17], the additional stage of chloroform-methanol deproteinization at the ratio 24:1 is preceded by the stage of adding 1.1 volume of CTAB to the double centrifugation supernatant.
The selected aqueous phase is precipitated by 2-propanol in all the three methods. The subsequent deposited DNA washing in the Savazzini & Martinelli method [14] is performed with 70% ethanol, while in the Pereira method [16] with a washing buffer containing ethanol and ammonium acetate, and in the Bigliazzi method [17] this stage is unavailable.
However, after the precipitated DNA resuspension with an eluent, the Bigliazzi method [17] incorporates additional resuspended DNA treatment with proteinase K, and subsequent manipulations, including the use of QIAprep Spin Miniprep Kit.
It must be noted that a number of commercial kits selectively binding extractable nucleic acid on spin columns, such as “Plant Genomic DNA Miniprep Kit” (Sigma), “Dneasy Plant Mini Kit” (Qiagen), “High Pure PCR Template Preparation Kit” (Roche), “Power Plant DNA Isolation Kit” (MO-BIO), “Power Soil DNA Isolation Kit” (MO-BIO), “FastDNA Spin Kit for Soil” (MP Biomedicals), “NucleoSpin Food” (Macherey-Nagel), “Genomic DNA from Food” (Macherey-Nagel), and “Dneasy Mericon Food Kit” (Qiagen) were tested in Vitis vinifera L. DNA extraction, including the use of “DNA Purification System for Food” (Promega) based on DNA extraction with the help of magnetic particles [11].
At the same time, the procedure of DNA isolation by commercial sets from the 2-isopropanol precipitated plant debris had minor modifications associated with the introduction of a set of 100 µl α-amylase (incubation at 80°C for 30 min) and/or 40 µl of proteinase K (incubation at 55°C for 30 min) into the first used buffer [11].
Sequential application of α-amylase and proteinase K is also provided by the procedure of nucleic acid isolation from lyophilized, pre-dissolved in a buffer (0.1 M Tris-HCl (pH 8.0), and 0.1 M NaCl) wine powder whose DNA extraction protocol is described in Nakamura et al. (2007) [15]. In this case, proteinase K is included in the hydrolysis by incubation at 55°C for 60 min together with sodium dodecyl sulfate (SDS). The resulting hydrolysate undergoes stepwise stages of precipitation by centrifugation in a mixture with 2-propanol; nucleic acid precipitate resuspension with an eluent buffer followed by the addition of 70% ethanol; re-deposition in a mixture of 2-propanol with sodium acetate; and deposited DNA resuspension in TE buffer. The extraction procedure also includes the stages of RNA removal by treatment of nucleic acid with RNase A and deproteinization first with neutral phenol and then with phenol:chloroform (1:1), with precipitation of the selected aqueous phase by addition of 0.2 M NaCl and 2 volumes of cold ethanol. The resulting DNA precipitate is washed with 70% ethanol and eluted with TE buffer.
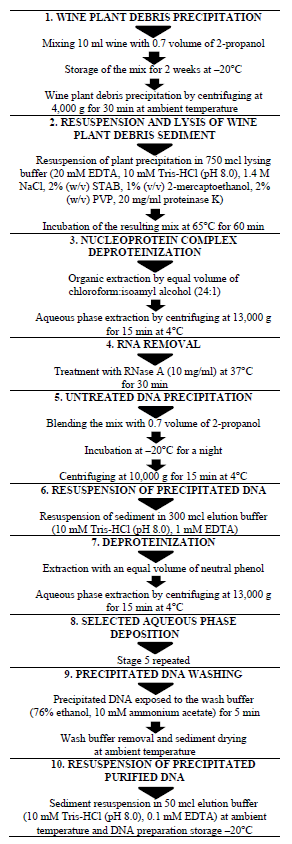
Performance analysis of Vitis vinifera L. DNA extraction by different methods indicates the superiority of the Pereira method [16] over other traditional methods of extraction [13–15, 17] in terms of DNA yield and quality [11]. Detailed information on the stages of Vitis vinifera L. sample preparation and DNA isolation by the Pereira method [16] is shown in Fig. 1.
The comparative quantitative and qualitative assessment of the isolated nucleic acid preparations pointed to the ineffectiveness of the Bigliazzi method [17]. A number of commercial sets with the exception of “Dneasy Plant Mini Kit” (Qiagen), “High Pure PCR Template Preparation Kit” (Roche), and “Power Soil Isolation Kit” (MO-BIO) with their minor modifications associated with the addition of α-amylase and/or proteinase K providing test systems performance, also proved ineffective. Treatment with α-amylase is prescribed for the set produced by Roche, while for the MO-BIO set [11] it is α-amylase and proteinase K treatment.
Precipitators used for wine plant debris precipitation, such as NaCl, 2-propanol, sodium acetate used both individually and in a complex are the key reagents at the initial stage of sample preparation which involves concentration of the test material by centrifugation. From the point of view of the efficiency of DNA yield and quality, the method is not inferior to concentration by lyophilization though rather time-consuming. For example, the Pereira method [16] regulates storage of wine and precipitator mixture in the freezer for 2 weeks for maximum precipitation effect.
Extracted from wines Vitis vinifera L DNA has the status of residual nucleic acid since its concentration is significantly reduced during the multi-stage wine production process including decantation, purification, filtration and other processing methods [15, 20]. In addition, grapes DNA is degraded by the DNase of wine microbiota during fermentation [11].
Wine aging reduces the yield of the extracted nucleic acid [22]. At the same time, it is experimentally proved [11] that the amount of Vitis vinifera L DNA isolated from wines with the completed stage of alcoholic fermentation is significantly reduced or practically absent depending on the terms of end product testing.
Methods of wines DNA authentication. The use of highly polymorphic microsatellite DNA loci designed for grapes genetic identification [23–27] is one of the approaches to wine DNA authentication as well [11, 13–17, 21, 22, 28, 29, 30].
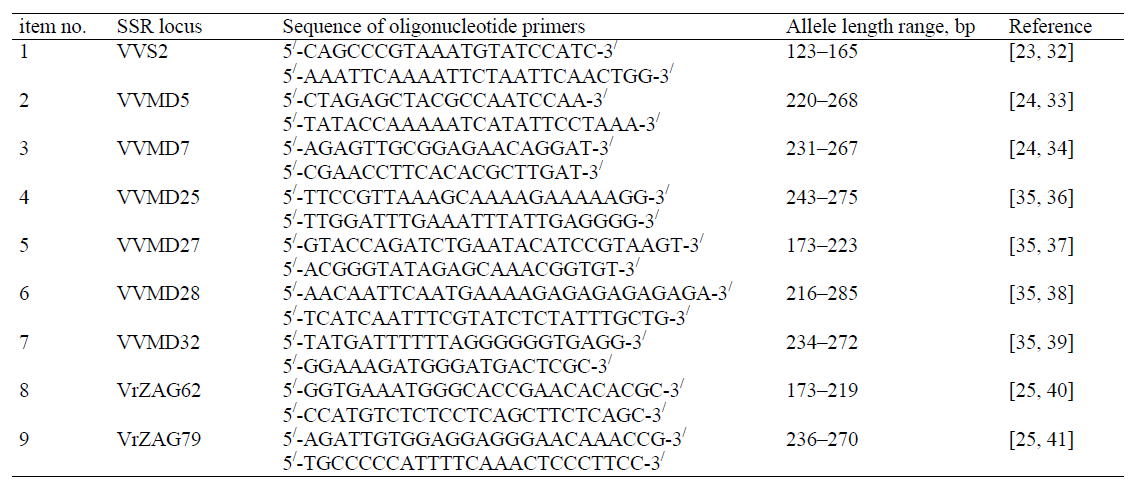
Nine highly specific and reproducible polymorphic markers of the nuclear DNA microsatellite (SSR) loci presented in Table 2 form the basic set for identification and certification of grapes varieties and hybrids, where the specified sequences of oligonucleotide primers initiate amplification of SSR fragments of the extracted DNA for subsequent granular analysis in capillary gel electrophoresis [31] of the sequencer with interpretation of results by the genetic analyzer software.
SSR fragments amplification is carried out by multiplex PCR, combining several analyzed loci. This amplification strategy is based on working with DNA extracted from the components of grape plants (fruit, leaf, stem, root) but is not effective while studying extracted residual wine nucleic acid [11, 13, 16, 42]. Therefore, wine DNA authentication is usually carried out by PCR with a set of primers of a single SSR- marker to achieve an analyzable result [11].
While DNA testing monosort and assemblage commercial wines researchers noted identifiable results of the markers specified in Table 2: VrZAG79 [11, 16], VVS2, VVMD27 [14, 17], and VVMD25 [17] These allow for a retrospective assessment of the wine materials varietal identity and actual wine DNA authentication by the interpretation of the fragmented data analysis.
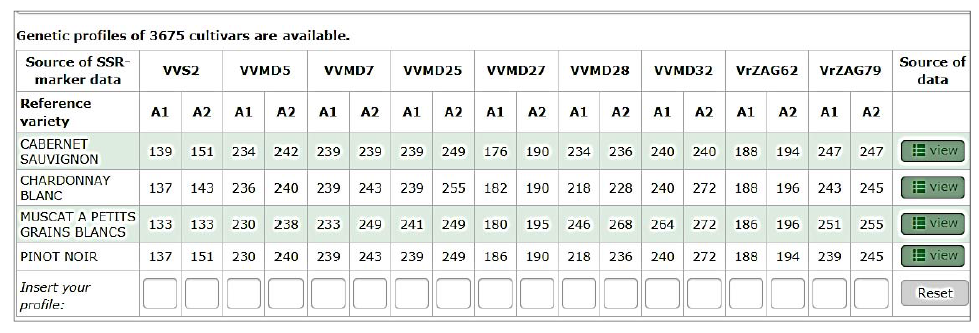
The database of 3675 grape varieties genetic profiles in VIVC interactive catalogue (Vitis International Variety Catalogue) (Fig. 2) is successfully used when comparing the DNA-test-generated SSR markers profiles with the already hosted on the server published information.
Another type of SSR markers, spSSR [43–46], targeted to the chloroplast DNA, has several advantages over the nuclear DNA (nSSR) analysis due to the greater number of representation per cell, greater resistance to exonuclease influence and lower susceptibility to degradation due to its content in double membrane organelles [11, 13].
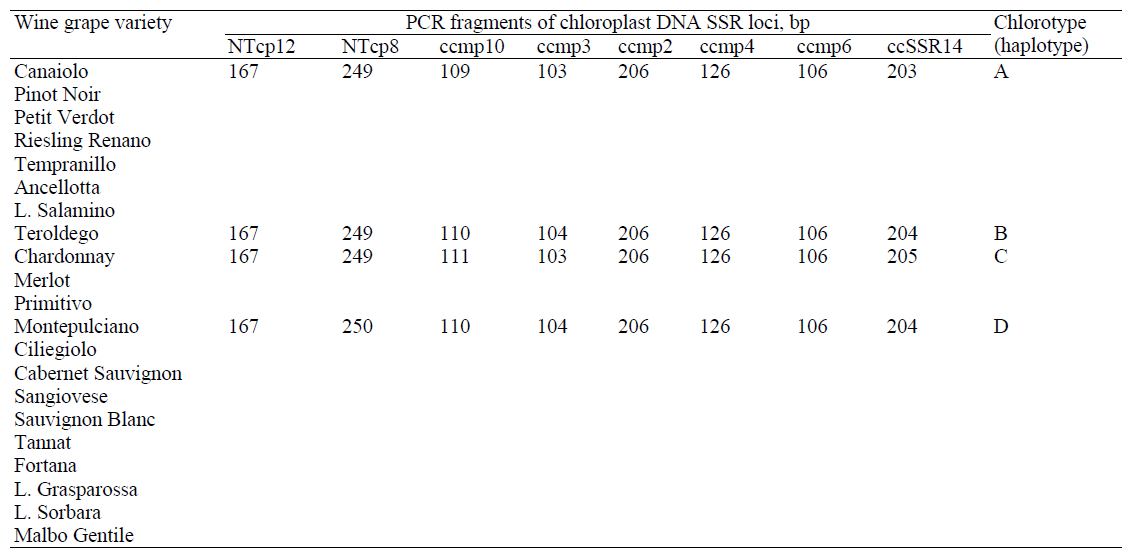
Despite the weak discriminatory ability of these SSR markers, incapable of wide range of grape varieties certification, the analysis of chloroplast DNA microsatellite loci remains an alternative approach to the varietal genetic identification of Vitis vinifera L, although not quite suitable for wine DNA authentication [11, 13, 16, 20, 21, 29] due to the low level of polymorphism of the analyzed loci, suitable only for a limited range of wine products differentiation (Table 3).
In the study of 21 grape varieties by fragmented analysis of 8 cpSSR-loci, whose oligonucleotide primers are listed in Table. 3, in the studied sample selection V. Catalano et al. (2016) [11] discovered 4 chlorotypes, whose results with grouping of the tested Vitis vinifera L varieties by their haplotype are shown in Table 4.
At the same time, half of these cpSSR-loci (NTcp12, ccmp2, ccmp4, and ccmp6) had no allele polymorphism, but two analyzed loci (ccmp10 and ccSSR14) were characterized by the presence of three alleles, and two more loci (NTcp8 and ccmp3) – by the presence of two alleles [11], respectively (Table 4).
Although this method shows relatively low resolution [11], it can be used as an additional test for counterfeit and adulterated wine products identification.
Microsatellite DNA is also used as a source of STS (Sequence Tagged Site – sites marked with a sequence) – unbroken unique sequences whose amplified profiles serve as molecular genetic markers [11, 15, 23]. Thus, S. Nakamura et al. (2007) [15] designed experimental sets of STS primers for certain SSR loci of mitochondrial and chloroplast DNA [25, 43, 51–54], having tested them in PCR during varietal genetic identification of Vitis vinifera L, and DNA authentication of wines produced from them.
Along with SSR markers, SNP markers [55, 56], applicable for wine DNA authentication [12], both thanks to tracing Vitis vinifera L. individual genotypes in monosort and assemblage wines with the potential for the quantitative assessment of plant ingredients and their performance in the analysis of the fragmented nucleic acid bear a high identification potential. Based on SNP markers, test systems can be designed for genetic identification of individual grape varieties [11].
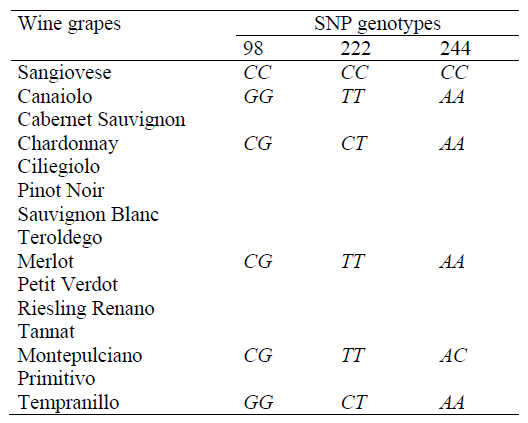
Table 5 shows the sets of primers and probes for real-time PCR with fluorescent hybridization detection used in genetic identification of the Sangiovese variety, and DNA authentication of the wine produced from it by assessing single-nucleotide polymorphism (SNP) in three analytical positions (98, 222 and 244) [11].
Sangiovese variety genetic identification and DNA authentication of the wine produced from it, based on the assessment of single-nucleotide polymorphism is established by the presence of allele C in the homozygous state (CC genotype) in all the three SNP analytical positions [11] (Table 6).
Another type of SNP markers application is the use of knowledge about single nucleotide polymorphism in a number of Vitis vinifera L genes integrated into melting curves analysis with high resolution (HRM analysis) on PCR platforms in real time [12, 57, 58].
HRM analysis is an effective technology of genotyping [59] with combined stages of PCR and detection of a high degree of specificity and sensitivity, capable of differentiating between several genotypes within one analysis, and suitable for wine DNA authentication [12, 57].
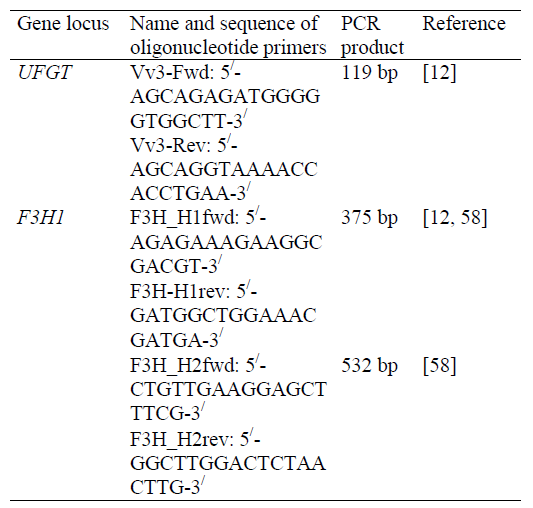
Table 7 presents the sets of PCR primers with subsequent HRM analysis and/or sequencing by grape varieties genetic identification and DNA authentication of the wines produced from them.
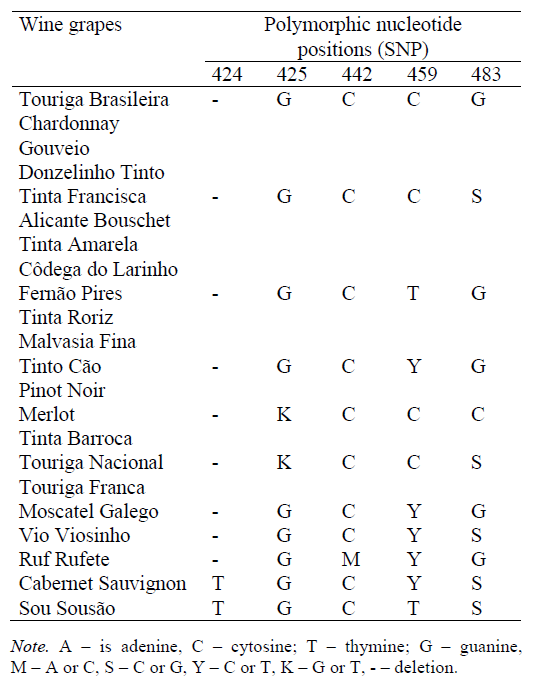
Primers Vv3-Fwd and Vv3-Rev initiate amplification of the UFGT gene locus, 119 bp long, with localization of the flanked region in the range of 387–505, covering five SNP analytical positions (424, 425, 442, 459, 483) [12] interpreted by HRM analysis and/or sequencing (Table 8).
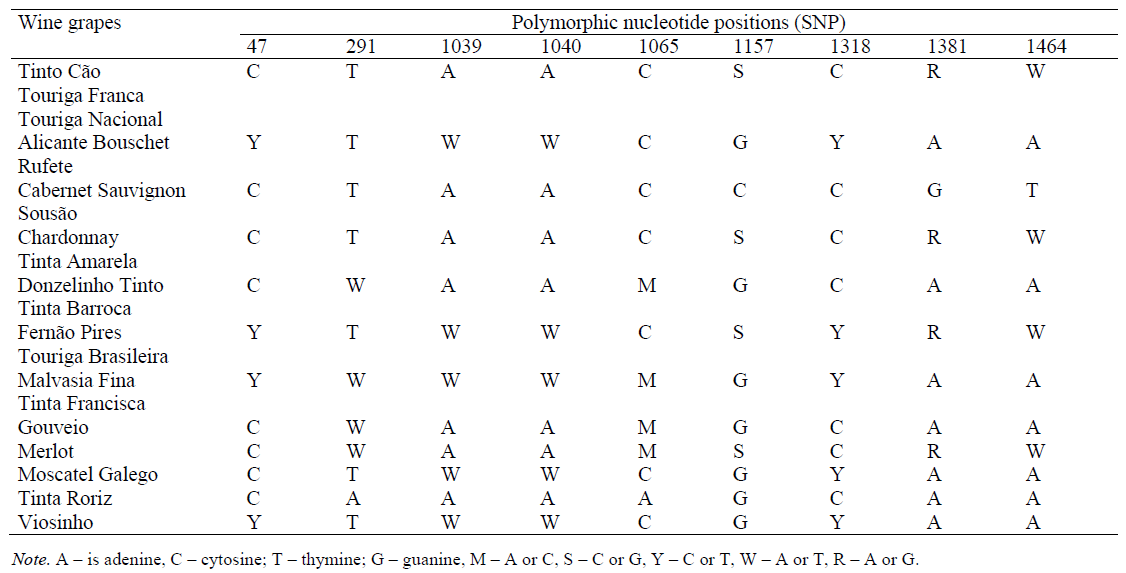
F3H_H1fwd and F3H-H1rev primers initiate amplification of the F3H1 gene locus, 375 bp long, with localization of the flanked region in the range of 5–379, covering two SNP analytical positions (47 and 291) [12, 58]; F3H_H2fwd and F3H_H2rev primers initiate amplification of the F3H1 gene locus, 532 bp long [30], with the flanked region localization in the range of 975–1506, covering eight SNP analytical positions (1039, 1040, 1065, 1157, 1318, 1381 and 1464) [58], respectively, also interpreted by HRM analysis and/or sequencing (Table 9).
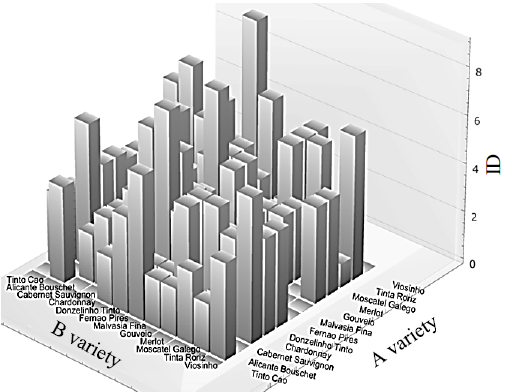
As can be seen from the considered examples, single gene locus SNP identification does not allow the origin of the analyzed wine material, to be established unambiguously therefore, it is not universal. In order to be able to determine the effectiveness of identification methods, as well as their combinations, the concept of identification distance was introduced for the first time. The identification distance (ID) of grape variety A from grape variety B is the number of polymorphic nucleotide positions (SNP) which allow identifying the presence of grape variety B DNA material impurities in wine material A. As seen from the definition, ID is asymmetric due to the presence of mixed nucleotides. For example, F3H-gene locus SNP analysis (Table 9) allows to determine the presence of Chardonnay grape variety in Cabernet Sauvignon wine material while detecting polymorphic nucleotide positions 1157, 1381 and 1464 (ID = 3). At the same time, it is not possible to differentiate Cabernet Sauvignon grape variety in Chardonnay wine material by this approach (ID = 0).
Fig. 3 shows the diagram of identification distances according to F3H-gene locus SNP analysis (Table 9), where the varieties with identical polymorphic nucleotide positions are combined into a single group.
Thus, it is urgent to create a complex method of wine DNA authentication, allowing a robust analysis procedure in the context of identification distance (namely, uncovering within the method the ID minimum threshold value for all variety pairs that is acceptable for the purposes of standardization) to identify and differentiate grape varieties in both varietal and assembling commercial wines
The arsenal of molecular genetic markers used for wine DNA authentication can be significantly expanded by the development of genomic and postgenomic technologies, with the introduction of experimental developments in the product quality management system based on the standards developed for the wine industry, especially while identifying wines of protected geographical indications, wines of protected designations of origin, and for the purposes of counterfeit products identification [1, 2, 60].
In the meantime, the existing methods of wine DNA authentication, published in scientific literature, have a recommendatory status, are not regulated by GOST/ISO and other legal documents of the Russian Federation, the Customs Union, the International Organization of Vine and Wine, and the European Union.
Legal and regulatory replenishment of the approved wine identification complex scheme with new DNA authentication methods will increase assessment reliability of authenticity and place of origin of wine products.
ВЫВОДЫ
Analysis of research and methodological approaches to the extraction of residual amounts of nucleic acids in wine raw materials and wine DNA authentication confirms the relevance of this research line and the prospects of its integration into the system of monitoring counterfeit and adulterated wine products. It shows the possibility of determining wine products authenticity and place of origin by DNA technologies, whose use implies ensured traceability of the product’s entire life cycle. The concept of identification distance (ID) between grape varieties A and B is introduced. ID stands for the number of polymorphous nucleotide positions (SNP), which allow the presence of grape variety B DNA material in variety A wine material to be identified. The algorithm of ID calculation and analysis of the information obtained is offered.
КОНФЛИКТ ИНТЕРЕСОВ
The authors declare no conflict of interest.
ФИНАНСИРОВАНИЕ
The study was supported by the section of agricultural products storage and processing of the Department of Agricultural Sciences of RAS and ‘V.M. Gorbatov FSC Foods Systems’ of RAS.
СПИСОК ЛИТЕРАТУРЫ
- Oganesyants L.A. Falʹsifikaty vinodelʹcheskoy produktsii: metody vyyavleniya [Counterfeit wine products: detection methods]. Production Quality Control, 2017, no. 7, pp. 8–11. (In Russ.).
- Parkhomenko A.I. Identification and detection of wine falsifications for customs purposes. Obrazovanie i nauka bez granits: sotsialʹno-gumanitarnye nauki [Education and Science Without Borders: Social and Human Sciences], 2016, no. 3, pp. 298–301. (In Russ.).
- Ebeler S.E., Takeoka G.R., and Winterhalter P. Progress in Authentication of Food and Wine. ACS Symposium Series, 2011, vol. 1081. DOI: https://doi.org/10.1021/bk-2011-1081.
- Montet D. and Ray R.C. Food Traceability and Authenticity: Analytical Techniques. Boca Raton, Florida: CRC Press, 2017, 354 p. DOI: https://doi.org/10.1201/9781351228435.
- Anikina N.S., Gnilomedova N.V., Agafonova N.M., and Riabinina O.V. Peculiarities of regulatory requirements for the control of quality and safety of wines. Magarach. Viticulture and Enology, 2016, no. 3, pp. 37–43. (In Russ.).
- Yakuba Yu.F. and Temerdashev Z.A. Chromatography methods in the analysis and identification of grape wines. Analytics and Control, 2015, vol. 19, no. 4, pp. 288–301. DOI: https://doi.org/10.15826/analitika.2015.19.4.013. (In Russ.).
- Oganesyants L.A., Panasyuk A.L., Kuzmina E.I., and Kharlamova L.N. Determination of the carbon isotope 13C/12C in ethanol of fruit wines in order to define identification characteristics. Foods and Raw Materials, 2016, vol. 4, no. 1, pp. 141–147. DOI: https://doi.org/10.21179/2308-4057-2016-1-141-147.
- Christoph N., Hermann A., and Wachter H. 25 Years authentication of wine with stable isotope analysis in the European Union – Review and outlook. BIO Web of Conferences, 2015, no. 5. DOI: https://doi.org/10.1051/bioconf/20150502020.
- Oganesyants L.A., Khurshudyan S.A., and Galstyan A.G. Monitoring kachestva pishchevykh produktov – bazovyy ehlement strategii [Food quality monitoring as the basic strategic element]. Production Quality Control, 2018, no. 4, pp. 56–59. (In Russ.).
- İşçi B., Yildirim H.K., and Altindişli A. A Review of the Authentication of Wine Origin by Molecular Markers. Journal of the Institute of Brewing, 2009, vol. 115, no. 3, pp. 259–264. DOI: https://doi.org/10.1002/j.2050-0416.2009.tb00378.x.
- Catalano V., Moreno-Sanz P., Lorenzi S., and Grando M.S. Experimental Review of DNA-Based Methods for Wine Traceability and Development of a Single-Nucleotide Polymorphism (SNP) Genotyping Assay for Quantitative Varietal Authentication. Journal of Agricultural and Food Chemistry, 2016, vol. 64, no. 37, pp. 6969–6984. DOI: https://doi.org/10.1021/acs.jafc.6b02560.
- Pereira L., Gomes S., Castro C., et al. High Resolution Melting (HRM) applied to wine authenticity. Food Chemistry, 2017, vol. 216, pp. 80–86. DOI: https://doi.org/10.1016/j.foodchem.2016.07.185.
- Baleiras-Couto M.M. and Eiras-Dias J.E. Detection and identification of grape varieties in must and wine using nuclear and chloroplast microsatellite markers. Analytica Chimica Acta, 2006, vol. 563, no. 1–2, pp. 283–291. DOI: https://doi.org/10.1016/j.aca.2005.09.076.
- Savazzini F. and Martinelli L. DNA analysis in wines: Development of methods for enhanced extraction and real-time polymerase chain reaction quantification. Analytica Chimica Acta, 2006, vol. 536, no. 1–2, pp. 274–282. DOI: https://doi.org/10.1016/j.aca.2005.10.078.
- Nakamura S., Haraguchi K., Mitani N., and Ohtsubo K. Novel Preparation Method of Template DNAs from Wine for PCR to Differentiate Grape (Vitis vinifera L.) Cultivar. Journal of Agricultural and Food Chemistry, 2007, vol. 55, no. 25, pp. 10388–10395. DOI: https://doi.org/10.1021/jf072407u.
- Pereira L., Guedes-Pinto H., and Martins-Lopes P. An Enhanced Method for Vitis vinifera L. DNA Extraction from Wines. American Journal of Enology and Viticulture, 2011, vol. 62, no. 4, pp. 547–552. DOI: https://doi.org/10.5344/ajev.2011.10022.
- Bigliazzi J., Scali M., Paolucci E., Cresti M., and Vignani R. DNA Extracted with Optimized Protocols Can Be Genotyped to Reconstruct the Varietal Composition of Monovarietal Wines. American Journal of Enology and Viticulture, 2012, vol. 63, no. 4, pp. 568–573. DOI: https://doi.org/10.5344/ajev.2012.12014.
- Rodríguez-Plaza P., González R., Moreno-Arribas M.V., et al. Combining microsatellite markers and capillary gel electrophoresis with laser-induced fluorescence to identify the grape (Vitis vinifera) variety of musts. European Food Research and Technology, 2006, vol. 223, no. 5, pp. 625–631. DOI: https://doi.org/10.1007/s00217-005-0244-2.
- Drábek J., Stávek J., Jalvková M., Jurcek T., and Frébort I. Quantification of DNA during winemaking by fluorimetry and Vitis vinifera L.-specific quantitative PCR. European Food Research and Technology, 2008, vol. 226, no. 3, pp. 491–497. DOI: https://doi.org/10.1007/s00217-007-0561-8.
- Garcia-Beneytez E., Maria V.M., Joaquin B., Maria C.P., and Javier I. Application of a DNA Analysis Method for the Cultivar Identification of Grape Musts and Experimental and Commercial Wines of Vitis vinifera L. Using Microsatellite Markers. Journal of Agricultural and Food Chemistry, 2002, vol. 50, no. 21, pp. 6090–6096. DOI: https://doi.org/10.1021/jf0202077.
- Siret R., Gigaud O., Rosec J.P., and This P. Analysis of Grape Vitis vinifera L. DNA in Must Mixtures and Experimental Mixed Wines Using Microsatellite Markers. Journal of Agricultural and Food Chemistry, 2002, vol. 50, no. 13, pp. 3822–3827. DOI: https://doi.org/10.1021/jf011462e.
- Hârta M.H., Pamfil D., Pop R., and Vicaş S. DNA Fingerprinting Used for Testing Some Romanian Wine Varieties. Bulletin UASVM Horticulture, 2011, vol. 68, no. 1, pp. 143–148. DOI: https://doi.org/10.15835/buasvmcn-hort:7041.
- Thomas M.R. and Scott N.S. Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as sequence-tagged sites (STSs). Theoretical and Applied Genetics, 1993, vol. 86, no. 8, pp. 985–990. DOI: https://doi.org/10.1007/BF00211051.
- Bowers J.E., Dangl G.S., Vignani R., and Meredith C.P. Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome, 1996, vol. 39, no. 4, pp. 628–633. DOI: https://doi.org/10.1139/g96-080.
- Sefc K.M., Regner F., Turetschek E., Glössl J., and Steinkellner H. Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome, 1999, vol. 42, no. 3, pp. 367–373.
- Maul E., Töpfer R., Carka F., et al. Identification and characterization of grapevine genetic resources maintained in Eastern European Collections. Vitis - Journal of Grapevine Research, 2015, vol. 54, pp. 5–12.
- This P., Jung A., Boccacci P., et al. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theoretical and Applied Genetics, 2004, vol. 109, no. 7, pp. 1448–1458. DOI: https://doi.org/10.1007/s00122-004-1760-3.
- Siret R., Boursiquot J.M., Merle M.H., Cabanis J.C., and This P. Toward the Authentication of Varietal Wines by the Analysis of Grape (Vitis vinifera L.) Residual DNA in Must and Wine Using Microsatellite Markers. Journal of Agricultural and Food Chemistry, 2000, vol. 48, no. 10, pp. 5035–5040. DOI: https://doi.org/10.1021/jf991168a.
- Boccacci P., Akkak A., Marinoni D.T., Gerbi V., and Schneider A. Genetic traceability of Asti Spumante and Moscato d’Asti musts and wines using nuclear and chloroplast microsatellite markers. European Food Research and Technology, 2012, vol. 235, no. 3, pp. 439–446. DOI: https://doi.org/10.1007/s00217-012-1770-3.
- Pereira L., Martins-Lopes P., Batista C., et al. Molecular Markers for Assessing Must Varietal Origin. Food Analytical Methods, 2012, vol. 5, no. 6, pp. 1252–1259. DOI: https://doi.org/10.1007/s12161-012-9369-7.
- Sefc K.M., Lefort F., Grando M.S., et al. Microsatellite markers for grapevine: A state of the art. In: Roubelakis-Angelakis K.A. (ed) Molecular Biology and Biotechnology of the Grapevine. Dordrecht: Springer Netherlands Publ., 2001, pp. 433–463. DOI: https://doi.org/10.1007/978-94-017-2308-4.
- Pellerone F.I., Edwards K.J., and Thomas M.R. Grapevine microsatellite repeats: Isolation, characterization and use for genotyping of grape germplasm from Southern Italy. Vitis, 2001, vol. 40, no. 4, pp. 179–186.
- Hârta M. and Pamfil D. Molecular Characterisation of Romanian Grapevine Cultivars Using Nuclear Microsatellite Markers. Bulletin UASVM Horticulture, 2013, vol. 70, no. 1, pp. 131–136.
- Ghetea L.G., Motoc R.M., Popescu C.F., Barbacar N., et al. Genetic profiling of nine grapevine cultivars from Romania, based on SSR markers. Romanian Biotechnological Letters, 2010, vol. 15, no. 1, pp. 116–124.
- Bowers J.E., Dangl G.S., and Meredith C.P. Development and characterization of additional microsatellite DNA markers for grape. American Journal of Enology and Viticulture, 1999, vol. 50, no. 3, pp. 243–246.
- Dokupilováa I., Šturdíka E., and Mihálik D. Characterization of vine varieties by SSR markers. Acta Chimica Slovaca, 2013, vol. 6, no. 2, pp. 227–234. DOI: https://doi.org/10.2478/acs-2013-0035.
- Sant'Ana G.C., Ferreira J.L., Rocha H.S., et al. Comparison of a retrotransposon-based marker with microsatellite markers for discriminating accessions of Vitis vinifera. Genetics and Molecular Research, 2012, vol. 11, no. 2, pp. 1507–1525. DOI: https://doi.org/10.4238/2012.May.21.8.
- Carimi F., Mercati F., De Michele R., et al. Intra-varietal genetic diversity of the grapevine (Vitis vinifera L.) cultivar ‘Nero d’Avola’ as revealed by microsatellite markers. Genetic Resources and Crop Evolution, 2011, vol. 58, no. 7, pp. 967–975. DOI: https://doi.org/10.1007/s10722-011-9731-4.
- Guo D.-L., Yu Y.-H., Xi F.-F., Shi Y.-Y., and Zhang G.-H. Histological and Molecular Characterization of Grape Early Ripening Bud Mutant. International Journal of Genomics, 2016, 7 p. DOI: https://doi.org/10.1155/2016/5620106.
- Aversano R., Basile B., Buonincontri M.P., et al. Dating the beginning of the Roman viticultural model in the Western Mediterranean: The case study of Chianti (Central Italy). PLoS One, 2017, vol. 12, no. 11. DOI: https://doi.org/10.1371/journal.pone.0186298.
- Lukyanov A.A., Bolshakov V.A., Ilnitskaya E.T. Creation of database and DNA-sertification of varieties of anapic ampelographic collection. Fruit growing and viticulture of South Russia, 2018, vol. 51, no. 3, pp. 49–58. DOI: https://doi.org/10.30679 / 2219-5335-2018-3-51-49-58. (In Russ.).
- Scali M., Elisa P., Jacopo B., Mauro C., and Vignani R. Vineyards genetic monitoring and Vernaccia di San Gimignano wine molecular fingerprinting. Advances in Bioscience and Biotechnology, 2014, vol. 5, no. 2, pp. 142–154. DOI: https://doi.org/10.4236/abb.2014.52018.
- Chung S.-M. and Staub J.E. The development and evaluation of consensus chloroplast primer pairs that possess highly variable sequence regions in a diverse array of plant taxa. Theoretical and Applied Genetics, 2003, vol. 107, no. 4, pp. 757–767. DOI: https://doi.org/10.1007/s00122-003-1311-3.
- Weising K. and Gardner R.C. A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome, 1999, vol. 42, no. 1, pp. 9–19. DOI: https://doi.org/10.1139/g98-104.
- Arroyo-Garcı´a R., Lefort F., de Andre´s M.T., et al. Chloroplast microsatellite polymorphisms in Vitis species. Genome, 2002, vol. 45, no. 6, pp. 1142–1149. DOI: https://doi.org/10.1139/g02-087.
- Ebert D. and Peakall R. Chloroplast simple sequence repeats (cpSSRs): technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Molecular Ecology Resources, 2009, vol. 9, no. 3, pp. 673–690. DOI: https://doi.org/10.1111/j.1755-0998.2008.02319.x.
- Bryan G.J., McNicoll J., Ramsay G., and Meyer R.C. Polymorphic simple sequence repeat markers in chloroplast genomes of Solanaceous plants. Theoretical and Applied Genetics, 1999, vol. 99, pp. 859–867. DOI: https://doi.org/10.1007/s001220051306.
- Arroyo-García R., Ruiz-García L., Bolling L., et al. Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Molecular Ecology, 2006, vol. 15, no. 12, pp. 3707–3714. DOI: https://doi.org/10.1111/j.1365-294X.2006.03049.x.
- Weising K., Winter P., Hüttel B., and Kahl G. Microsatellite Markers for Molecular Breeding. Journal of Crop Production, 1997, vol. 1, no. 1, pp. 113–143. DOI: https://doi.org/10.1300/J144v01n01_06.
- Pe´ros J.-P., Berger G., Portemont A., Boursiquot J.-M., and Lacombe T. Genetic variation and biogeography of the disjunct Vitis subg. Vitis (Vitaceae). Journal of Biogeography, 2011, vol. 38, no. 3, pp. 471–486. DOI: https://doi.org/10.1111/j.1365-2699.2010.02410.x.
- Nishikawa T., Vaughan D.A., and Kadowaki K. Phylogenetic analysis of Oryza species, based on simple sequence repeats and their flanking nucleotide sequences from the mitochondrial and chloroplast genomes. Theoretical and Applied Genetics, 2005, vol. 110, no. 4, pp. 696–705. DOI: https://doi.org/10.1007/s00122-004-1895-2.
- Bergman, C.J., Bhattacharya, K.R., Ohtsubo, K. Rice end-use quality analysis. In: Champagne E.T. (ed) Rice: Chemistry and Technology. 3 ed. St. Paul, USA: American Association of Cereal Chemists Publ., 2004. pp. 415–472.
- Ohtsubo K. and Nakamura S. Cultivar Identification of Rice (Oryza sativa L.) by Polymerase Chain Reaction Method and Its Application to Processed Rice Products. Journal of Agricultural and Food Chemistry, 2007, vol. 55, no. 4, pp. 1501–1509. DOI: https://doi.org/10.1021/jf062737z.
- Ohtsubo K., Nakamura S., Yoza K., and Shishido K. Identification of glutinous rice cultivars using rice cake as samples by the PCR method. Journal of the Japanese Society for Food Science and Technology, 2001, vol. 48, no. 4, pp. 306–310.
- Lijavetzky D., Cabezas J.A., Ibáñez A., Rodríguez V., and Martínez-Zapater J.M. High throughput SNP discovery and genotyping in grapevine (Vitis vinifera L.) by combining a re-sequencing approach and SNPlex technology. BMC Genomics, 2007, vol. 8, pp. 424. DOI: https://doi.org/10.1186/1471-2164-8-424.
- Cabezas J.A., Ibáñez J., Lijavetzky D., et al. A 48 SNP set for grapevine cultivar identification. BMC Plant Biology, 2011, vol. 11, pp. 153. DOI: https://doi.org/10.1186/1471-2229-11-153.
- Pereira L. and Martins-Lopes P. Vitis vinifera L. Single-Nucleotide Polymorphism Detection with High-Resolution Melting Analysis Based on the UDP-Glucose: Flavonoid 3-O-Glucosyltransferase Gene. Journal of Agricultural and Food Chemistry, 2015, vol. 63, no. 41, pp. 9165–9174. DOI: https://doi.org/10.1021/acs.jafc.5b03463.
- Gomes S., Castro C., Barrias S., et al. Alternative SNP detection platforms, HRM and biosensors, for varietal identification in Vitis vinifera L. using F3H and LDOX genes. Scientific Reports, 2018, vol. 8, no. 1, pp. 5850. DOI: https://doi.org/10.1038/s41598-018-24158-9.
- Druml B. and Cichna-Markl M. High resolution melting (HRM) analysis of DNA – Its role and potential in food analysis. Food Chemistry, 2014, vol. 158, pp. 245–254. DOI: https://doi.org/10.1016/j.foodchem.2014.02.111.
- Petrov A.N., Khanferyan R.A., and Galstyan A.G. Current aspects of counteraction of foodstuff's falsification. Problems of Nutrition, 2016, vol. 85, no. 5, pp. 86–92. (In Russ.).