Аннотация
Napa cabbage waste contains an organic component, cellulose, which can be utilised as an ingredient for cellulose-degrading enzyme production with the help of indigenous yeast. The aim of the research was to identify and characterise potential indigenous yeast isolated from napa cabbage waste, which has cellulose-degrading activity. Indigenous yeast were isolated and characterised using the RapID Yeast Plus System, then turbidity was used to determine the yeast total population. Indigenous yeast was grown at napa cabbage waste at 27, 37, and 40°C for three days, and cellulose-degrading activity was determined by the Dinitrosalicylic Acid (DNS) method. The potential yeast isolate with the highest cellulose-degrading activity was identified by a sequence analysis of the rRNA gene internal transcribed spacer (ITS) region with using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′- TCCTCCGCTTATTGATATGC-3′). The results were compared to the GenBank database using the Basic Local Alignment Search Tools/BLAST algorithm. Three species of indigenous yeast were isolated from napa cabbage waste (S2, S6, and S8). S8, incubated at 37ºC for three days, demonstrated the highest cellulose-degrading enzyme activity (1.188 U/mL), with the average activity of 0.684U/mL. Species identification results indicated that the S8 isolate had a 100% similarity to Pichia fermentans UniFGPF2 (KT029805.1).Ключевые слова
Pichia fermentans, temperature, cellulase enzyme, internal transcribed spacerВВЕДЕНИЕ
Napa cabbage (Brassica pekinesis L.) is one of the most cultivated agricultural products in Indonesia. In 2014 its production reached 602 468 t. The local leader in this field was the West Java province that yields 14.92 tons of napa cabbage per ha [1]. Since over 20% of napa cabbage cannot be utilised [2], this waste amount makes napa cabbage production inefficient.
Napa cabbage waste contains the same essential component, namely, polysaccharides in the form of cellulose, as napa cabbage itself. Cellulose is known to be a constituent component of plant cell walls, and it account for as much as 30–50% of total lignocellulose [3]. Currently, napa cabbage waste is used as animal feed, while its value could be increased, e.g., through production of cellulose-degrading enzymes.
Cellulose-degrading enzymes can be produced from napa cabbage waste, which is high in cellulose content, by yeast. Enzyme production by indigenous cellulolytic yeast requires optimal conditions, however, it is influenced by external factors, especially, temperature. Thus, too low temperatures can inhibit enzyme production because of the plasma membrane fluidity decrease which leads to disturbed metabolic activity [4]. On the other hand, too high temperature can damage cells and the structure of proteins, which are constituents of enzymes. The fact that temperature is an easily controlled parameter makes it possible to support yeast growth during the fermentation of napa cabbage waste for cellulose-degrading enzyme production. Therefore, the aim of this research was to characterise and identify indigenous yeast isolated from napa cabbage waste.
ОБЪЕКТЫ И МЕТОДЫ ИССЛЕДОВАНИЯ
The object of the research was napa cabbage waste from the Gedebage Central Market in Bandung City, Indonesia. We used the following materials: Potato Dextrose Agar (PDA), Yeast and Mold Agar (YMA), Nutrient Broth (NB), Thermo Scientific RapID Yeast Plus System Kit, Carboxymethyle Cellulose (CMC), distilled water, 0.85% NaCl, DNS reagent (3.5-Dinitrosalicyclic acid), phosphate buffer solution (pH 7), gelatin, antibiotics, KH2PO4, and MgSO4.
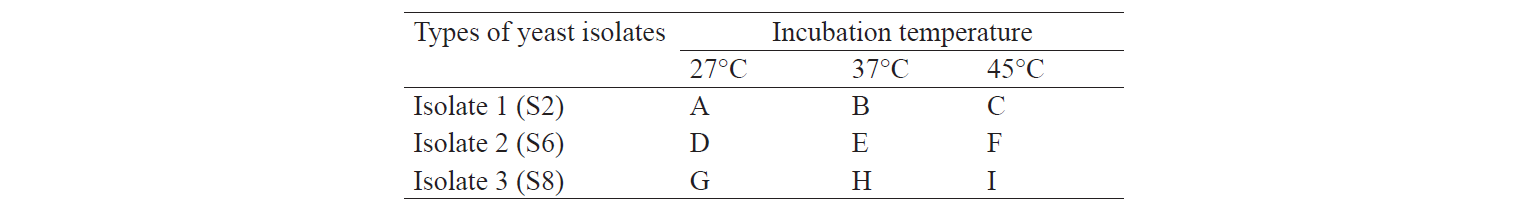
In our experiment we used nine treatments. Treatment factors were the type of yeast isolate and incubation temperature (Table 1). The isolation process of indigenous cellulolytic yeast from each treatment lasted for three days. The experiments were repeated three times.
The selection of the best treatment was performed based on quantitative analysis by determining the highest value of enzyme activity using Factorial Randomized Block Design. According to the results of isolation and identification of indigenous cellulolyticyeast from napa cabbage waste, descriptive analysis onthe total population of yeast during the production of cellulose-degrading enzymes was conducted.
Isolation and identification of indigenous yeast. The isolation of indigenous cellulolytic yeast from napa cabbage waste was carried out using the direct plating method [5–6]. One gram of crushed napa cabbage waste was added into 0.85% NaCl, inoculated into a modified PDA (PDA with a 3% yeast extract and 10 ppm antibiotics) and then incubated at 30°C for three days. The biochemical activities of the selected isolates were characterised with the help of RapID Yeast Plus System Kit [7]. For species identification of potential indigenous yeast with the highest cellulose-degrading enzyme activity we used rRNA gene internal transcribed spacer (ITS) region. Sequence analysis was carried out using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) as forward and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) as reverse. DNA amplification was performed by Macrogen Inc. The results were compared with the GenBank database using the BLAST algorithm [8].
Determination of total yeast population. Total yeast population determination was carried out by turbidimetry: 1 mL of the liquid culture was taken from the enzyme production medium followed by absorbance measurement [9]. This method is based on the spectrophotometric measurement of the total population at a wavelength of 600 nm [5].
Determination of cellulose-degrading enzyme activity. Cellulose-degrading enzyme production was carried out by the International Union’s recommended method of Pure and Applied Chemistry (IUPAC) with some modifications [10]. The salt media used consisted of KH2PO4, Mg2SO4 and gelatin. The napa cabbage waste was incubated in salt media at (1:2, w/v) by adding 2% (v/v) of isolates [11]. The isolation was carried out in an incubator at 27°C, 37°C and 45°C for three days followed by stirring at 100 rpm for 60 min at room temperature. Then every 24 h the fermented solution was separated using a centrifuge to obtain crude cellulose-degrading enzymes in supernatant, where crude enzymes reacted with DNS (Dinitrosalicylic Acid) reagent. Finally, spectrophotometric analysis was carried out to obtain absorbance values which were used to determine cellulose-degrading enzyme activity. The control used was 3 mL of DNS reagent that was diluted to 25 mL by distilled water.
РЕЗУЛЬТАТЫ И ИХ ОБСУЖДЕНИЕ
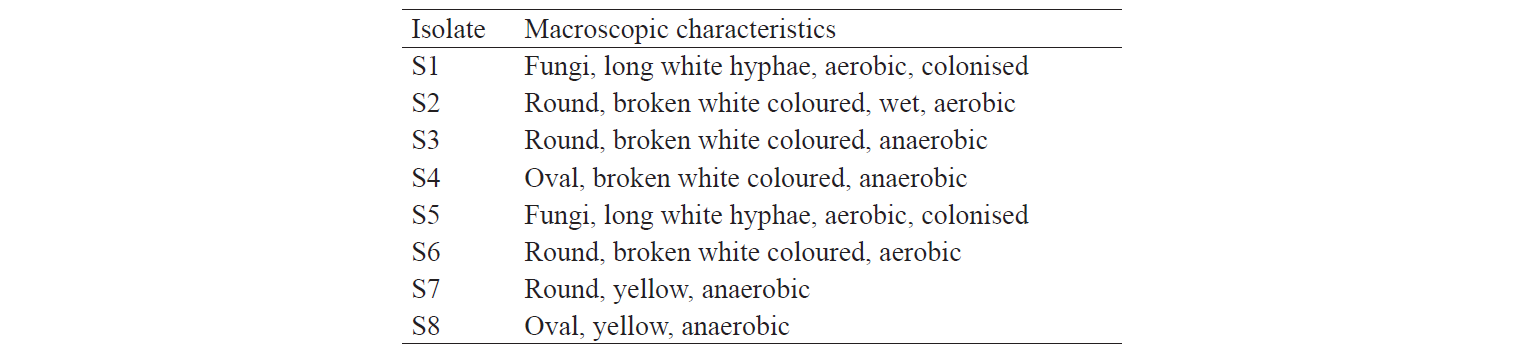
Characterisation of indigenous yeast. After three days of incubation, eight isolates with different characteristics were obtained (Table 2). S2, S6 and S8 isolates displayed macroscopic morphological characteristics similar to those of yeast.
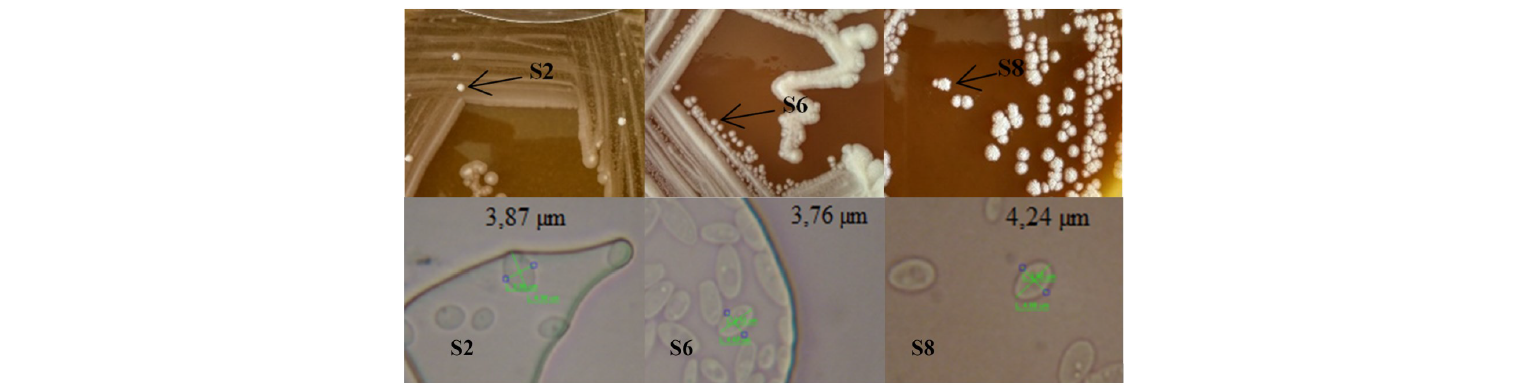
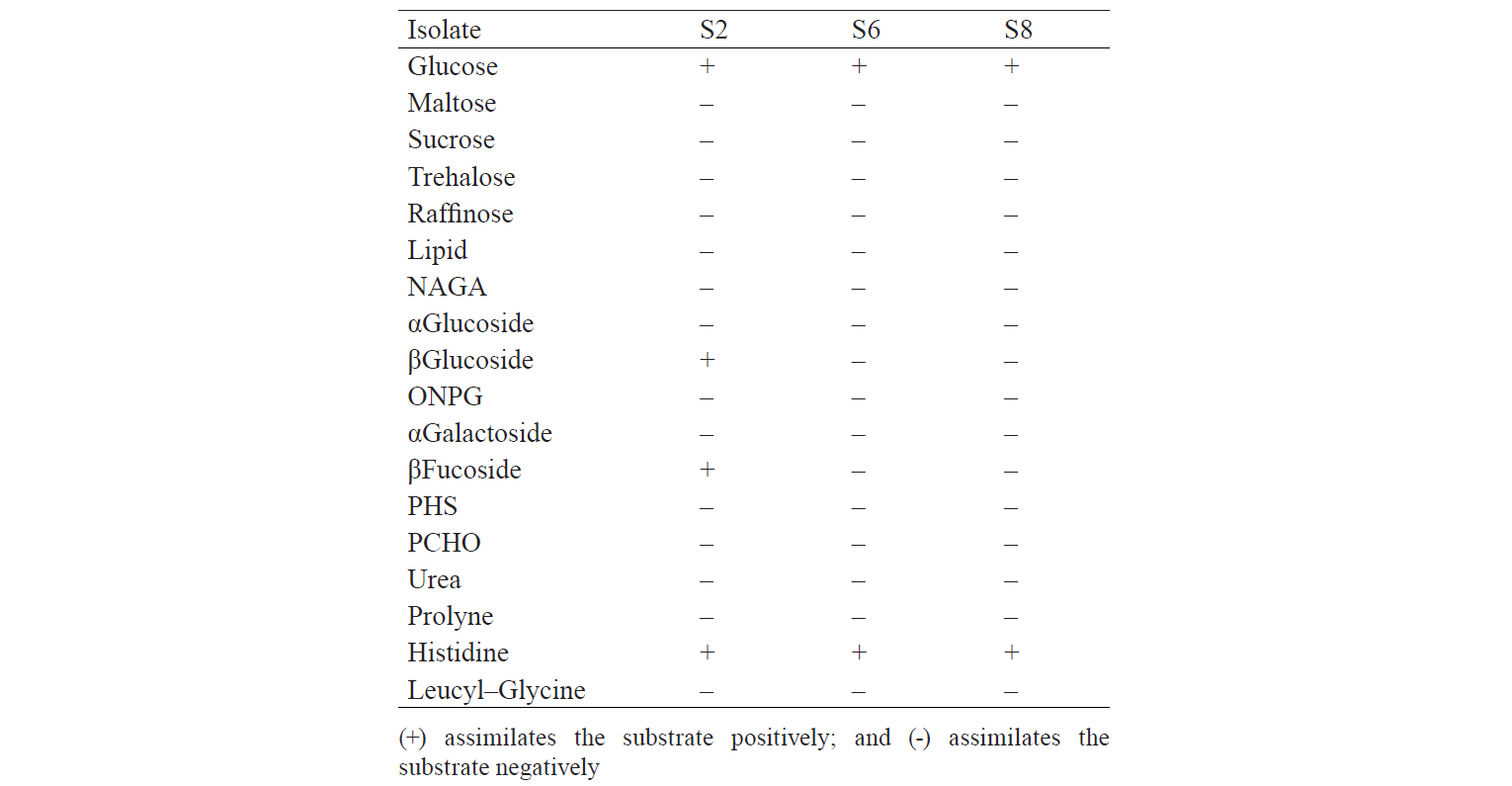
Asliha and Alami state that macroscopically, yeast are round, white, with membranous colony texture, while microscopically, multilateral yeast bud and its cell size ranges from 1 to 7 μm [12]. In addition, the microscopic analysis allowed three isolates to be chosen because they had cell size classified as that of yeast. An average cell diameter of the S2, S6 and S8 was 3.87, 3.76 and 4.24 μm, respectively (Fig. 1). The selected isolates were purified, and their biochemical activities were tested using the RapID Yeast Plus System (Table 3).
The identification results are based on the biochemical properties of the isolates tested against the reacted compound. The glucose or glucoside hydrolysis ability was only shown by S2 isolates against β-Glucoside and β-Fucoside. Lopez et al. states that several non-Saccharomyces yeast could be found in soil, fruits, trees, and damaged food or drink that has glycolytic β-glucosidase activity [13]. The biochemical properties of indigenous yeast (Table 5) are also supported by Mateo et al., who found the glycolytic activity, especially β-glucosidase activity, on indigenous yeas [14]. It implies that glucose can be hydrolysed into acidic compounds which reduce pH until it changes the colour of the resultants. Macroscopically, two isolates identified as S6 and S8 had different characteristics. They differed from the characteristics of Candida sp. that has the anamorphous properties, does not have a sexual reproduction phase, and has unstable phenotypic characteristics [15]. Therefore, although the two isolates were different in form, colour, and oxygen requirements, they had the same biochemical activity.
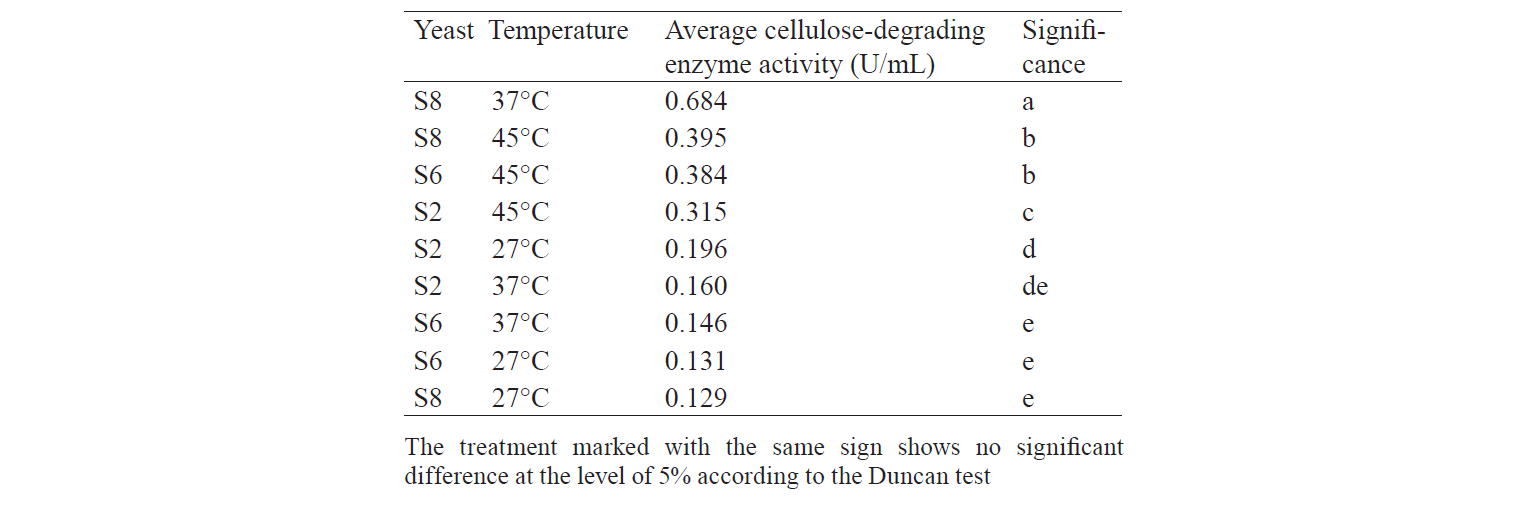
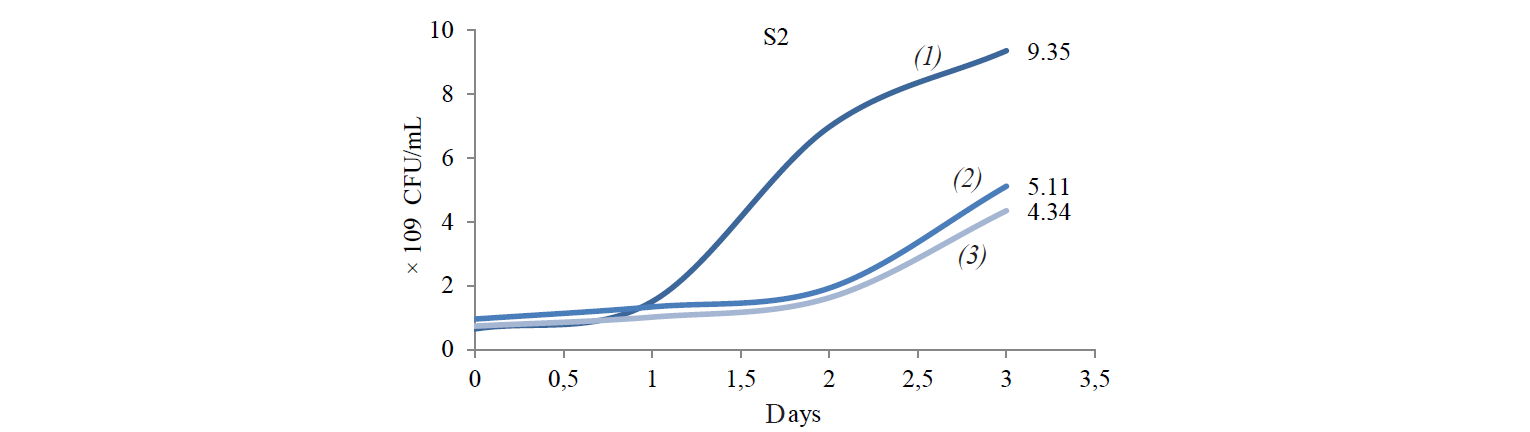
Total population of indigenous yeasts. The results of indigenous yeasts total population determination during incubation are demonstrated in Fig. 2. During day 1 of incubation, the total population in all treatments decreased because the isolates still were in the adaptation phase in the medium. This phase is called the lag phase or the cell adaptation period of new microorganisms to the environment [16]. Nguong et al. states that it takes 16 h for yeast with biochemical activity similar to that of the S2 isolate to adjust to a new environment [17].
After the adaptation phase, the total population of all treatments increased. The increase is the exponential growth phase, where cells of microorganisms have adapted to the environment and began to multiply so that the number of mass cells or cell density increases rapidly [16]. Spectrometric analysis preformed by Kanti et al. revealed that the population of indigenous yeast, such as Candida, Rhodothorula, Pichia, and Debaryomyces, began to increase from 24th h and reached a plateau by the 96th h (OD 600 nm) [5].
The highest total population was observed at the incubation temperature of 27°C in all the treatments. Mateo et al. state that the biochemical activity of the indigenous yeast was maximal at the temperature of 30–40°C [14]. However, the isolate that belongs to the Hanseniaspora genus that has biochemical activity similar to that of the S2 isolate had the maximum biochemical activity at 28°C. Meanwhile, according to Gänzle et al., a representative of the Candida genus with biochemical activity similar to that of S6 and S8 isolates grew rapidly at 27°C [18].
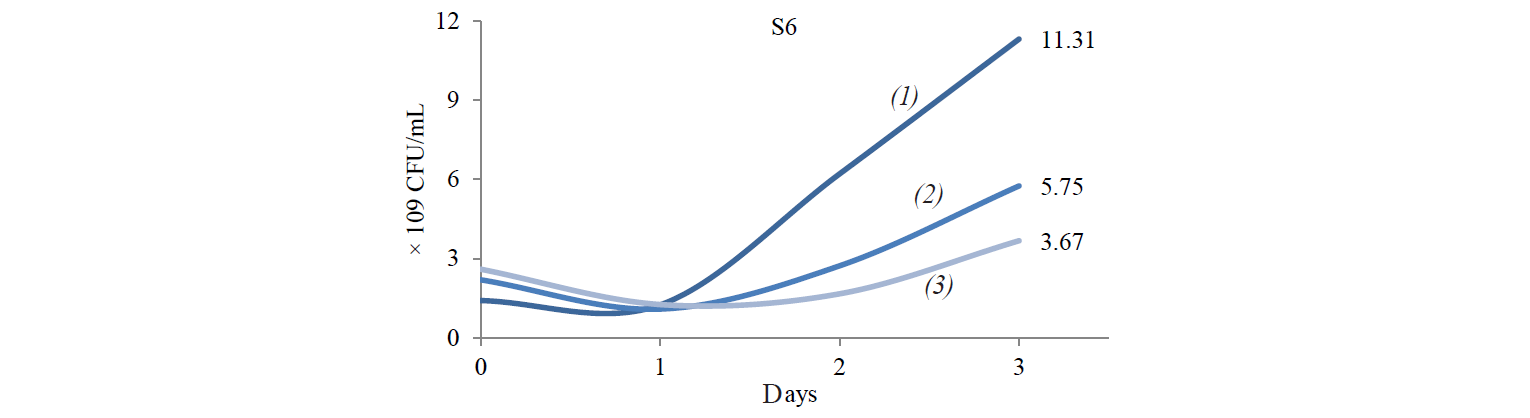
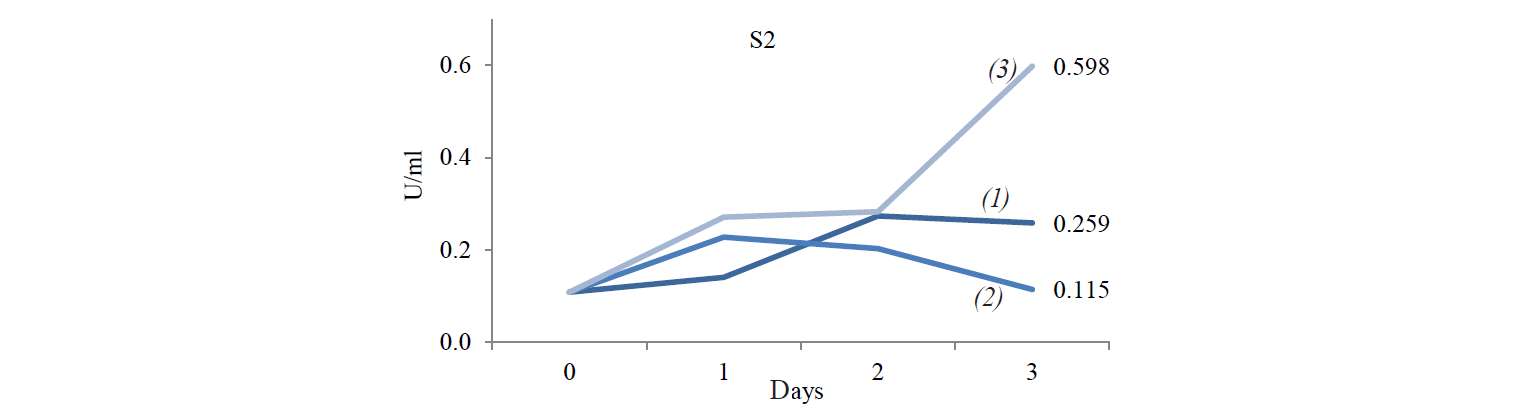
Cellulose-degrading enzyme activity. Cellulosedegrading enzyme activity of indigenous yeast is shown in Fig. 3. The highest enzyme activity produced by S2 was 0.598 U/mL at an incubation temperature of 45°C. The high temperature caused an increase in the rate of biochemical reactions, especially for indigenous yeast that has similar biochemical activity with Hanseniaspora. Fennema states that a high temperature affect various reactions [19]. The enzyme belongs to the group of mesozyme enzymes (in the range of 20–50°C) [20]. According to López et al., the glycolytic activity (β-glucosidase) of H.guilliermondii at 28°C is about 0.064–2.887 U/mL [13, 21].
S6 isolates obtained at the incubation temperature of 45°C also displayed a high enzyme activity. It is because the growth of Candida-like organisms occurred at the maximum temperature (40–45°C) [20]. As stated by Shuler and Kargi, enzymes are Growth-associated products, i.e. the growth of microorganisms is directly proportional to the product concentration [16]. However, the S8 isolate demonstrated the highest enzyme activity when treated at 37°C: its value was 1.203 U/mL on day1 and 1.188 U/mL on day 2.
Table 4 shows analysis of variance. F-value was greater than Pvalue probability (0.05), which indicated the presence of at least one treatment that significantly differed from the others. Hence, it required an additional test, namely, the Duncan Test.
Table 5 demonstrates the Duncan Test results. According to the data, the S8 treatment incubated at 37°C produced enzyme with an activity significantly differing from the other treatments. This is in accordance with the result of Sulman and Rehman, that Candida-like organisms are able to produce cellulose-degrading enzymes with the highest activity at 37°C [11]. The growth of isolates at 27°C cannot produce enzymes with high activity because energy supply from the environment is low, while at 45°C the growth of isolates is inhibited and the structure of the enzyme is denatured so that the activity is not optimal. Therefore, incubation at 37°C gives enough energy for isolates to grow without damaging the structure of the enzyme produced.
Temperature greatly influences the enzymatic activity and rigorous of yeast cell membranes, and higher temperature can shorten the exponential phase of yeast growth. In addition, higher temperature can cause denaturation of ribosomes and membrane fluidity problems. Thus, 30–35°C is the optimal temperature for yeast metabolism, including the enzymatic activity [22].
The difference in S2 and S8 enzyme activities was due to their different biochemical abilities. Lopez et al. found that Hanseniaspora sp., which is similar to the S2 isolate, was able to assimilate glycerol, galactose and sucrose, unlike with Candida sp., which is similar to S8 [21, 23].
The different activity of the enzyme produced by S6 and S8 could be caused by different phylogenetics between the two isolates. According to Birmeta et al., Candida sp. that was mentioned as C. krusei has close proximity to P. fermentans having certainly different biochemical ability than C. krusei [24]. P. fermentans has an anamorphic form, Candida lambica, but it is not uncommon to find C. lambica mis-identification as C. krusei caused by similar biochemical abilities of the yeast. Nevertheless, C. lambica is able to assimilate xylose, compared to C. krusei cannot [25]. Meanwhile, the ability of yeast to assimilate xylose has not been determined by the RapID Yeast Plus System method, so differences between C. krusei and C. lambica have not been identified.
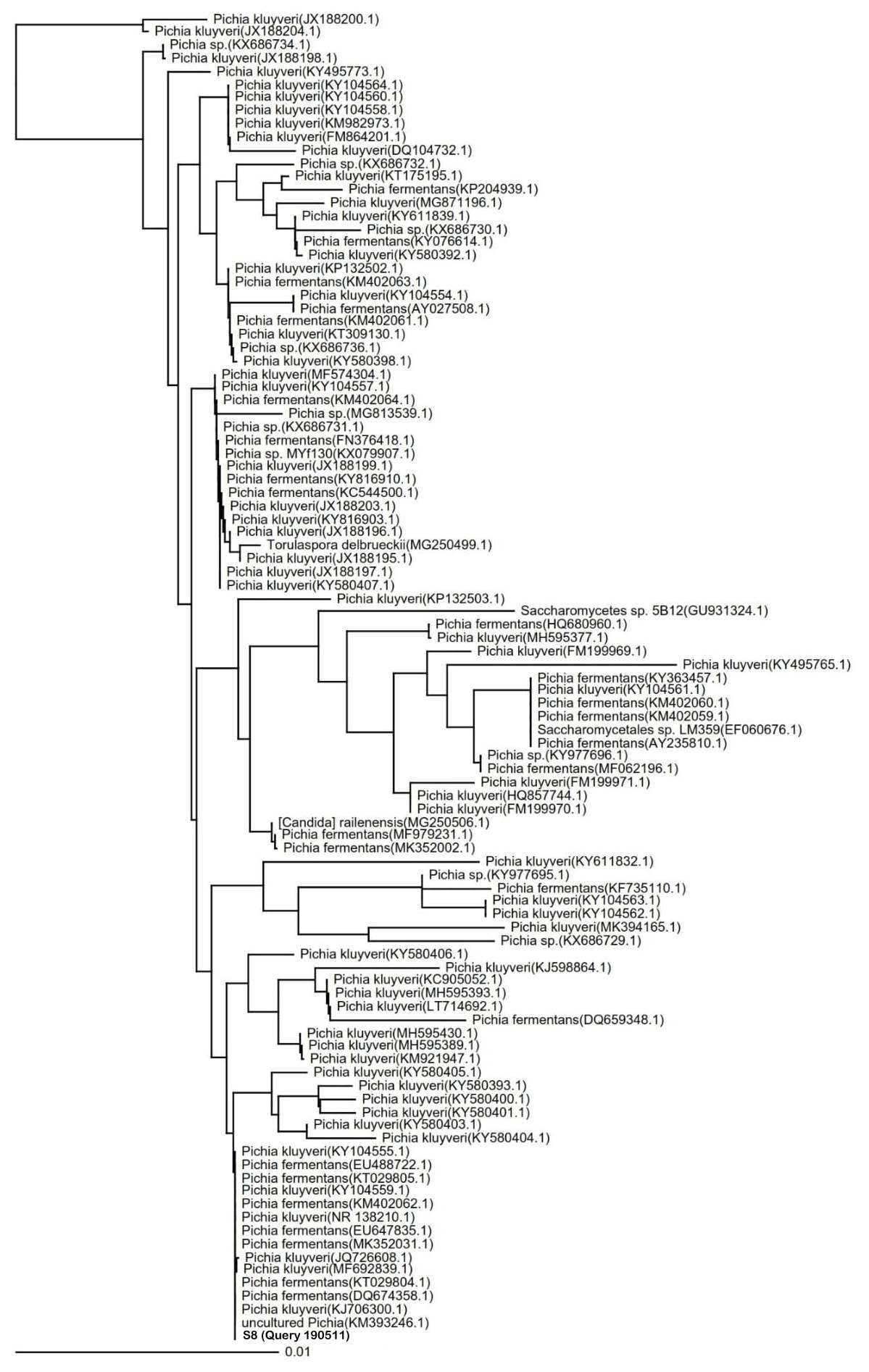
Species identification of potential indigenous yeast with the highest cellulose-degrading activities. The identification of the S8 isolate resulted in the 100% similarity to P. fermentans strain UniFGPF2 (KT029805.1). The phylogenetic tree (Fig. 4) shows that the S8 isolate is also similar to P. kluyveri culture CBS:188 (KY104555.1), P. fermentans strain UniFGPF1 (KT029804.1), P. fermentans strain UFLA CWFY24 (KM402062.1), and P. fermentans strain YF12b (EU488722.1, DQ674358.1).
P. fermentans have the ability to ferment and assimilate glucose, D-xylose, succinate, lactate, citrate, and glycerol [24]. Candida lambica is an anamorphic form of P. fermentans which can assimilate glucose and xylose but cannot assimilate arabinose, galactose, and selobiosa [26]. In addition, Issatchenkia orientalis, a teleomorphic form of Candida krusei that usually incorrectly identified as Candida lambica, can assimilate glucose sufficiently but cannot assimilate galactose, maltose, sucrose, lactose, raffinose, and trehalose [27].
According to Bengoa et al., despite P. fermentans and C. lambica can growth at a temperature up to 37°C, the optimal temperature is 25–30°C [28]. Such strain as I. orientalis has the unique properties, as this microorganism can grow at a higher temperature level. Miao et al. reported that I. orientalist strains optimally grows and produces a high amount of ethanol at 41°C, which indicates its thermostability [29].
ВЫВОДЫ
Three species of indigenous yeast were isolated from napa cabbage waste. The highest cellulosedegrading enzyme activity (1.188U/mL) displayed the S8 isolate incubated at 37°C for three days. Its average cellulose-degrading activity was 0.684U/mL. According To the species identification, the S8 isolate showed a 100% similarity to Pichia fermentans UniFGPF2 (KT029805.1).
КОНФЛИКТ ИНТЕРЕСОВ
The authors declare no conflict of interests.
БЛАГОДАРНОСТИ
The authors would like to thank the Student Research Group, Vivi Fadila Sari, Isfari Dinika and Syarah Virgina for their help with the experiments.
СПИСОК ЛИТЕРАТУРЫ
- Directorate General of Horticulture Statistik Produksi Hortikultura Tahun 2014. Kementerian pertanian: 2015. 315 p. (In Indonesian).
- Rahmah A, Izzati M, Parman S. The Effect of Liquid Organic Fertilizer Based on Basic Sawi Putih (Brassica chinensis L.) Waste on Growth of Sweet Corn Plant. Buletin Anatomi dan Fisiologi, 2014;22(1):65–71. DOI: https://doi.org/10.14710/baf.v22i1.7810.
- Saha BC. Lignocellulose Biodegradation and Applications in Biotechnology. In: Saha BC, Hayashi K, editors. Lignocellulose Biodegradation. American Chemical Society; 2004. pp. 2–34. DOI: https://doi.org/10.1021/bk-2004-0889.ch001.
- Beltran G, Rozes N, Mas A, Guillamon JM. Effect of low-temperature fermentation on yeast nitrogen metabolism. World Journal of Microbiology and Biotechnology. 2007;23(6):809–815. DOI: https://doi.org/10.1007/s11274-006-9302-6.
- Kanti A, Sudiana IM. Aktivitas CMC-ase khamir Candida sp. Yang diisolasi dari tanah kebun biologi wamena, Papua [CMC-ase Activity of Yeast Candida sp., Isolated from Soil of Wamena Biological Gardens, Papua]. Berita Biologi [Biology News]. 2003;6(5):655–660. (In Indonesian).
- Utama GL, Kurnani TBA, Sunardi, Balia RL. The Isolation and Identification of Stress Tolerance Ethanol-fermenting Yeasts from Mozzarella Cheese Whey. International Journal on Advanced Science, Engineering and Information Technology. 2016;6(2):252–257. DOI: http://doi.org/10.18517/ijaseit.6.2.752.
- Balia RL, Kurnani TBA, Utama GL. Selection of Mozzarella Cheese Whey Native Yeasts with Ethanol and Glucose Tolerance Ability. International Journal on Advanced Science, Engineering and Information Technology.2018;8(4):1091–1097. DOI:http://doi.org/10.18517/ijaseit.8.4.5869.
- Utama GL, Sidabutar FEE, Felina I, Wira DW, Balia RL. The utilization of fruit and vegetable wastes for bioethanol production with the inoculation of indigenous yeasts consortium. Bulgarian Journal of Agricultural Science. 2019;25(2):264–270.
- Diana L, Lasmini T. Isolasi dan Identifikasi Khamir Selulolitik Dari Tanah Rizosfer Anggrek Puser Bumi (Pecteilis susannae L.) di Hutan Wonosadi Gunung Kidul DIY [Isolation and Identification of Cellulolytic Yeast from Soil Rizosphere Puser Earth Orchid (Pecteilis susannae L.) in Wonosadi Forest Gunung Kidul DIY]. Biogenesis: Jurnal Ilmiah Biologi [Scientific Journal of Biology]. 2016;4(1):21–28. (In Indonesian). DOI: https://doi.org/10.24252/bio.v4i1.1116.
- Gupta P, Samant K, Sahu A. Isolation of Cellulose-Degrading Bacteria and Determination of Their Cellulolytic Potential. International Journal of Microbiology. 2012;2012. DOI: http://doi.org/10.1155/2012/578925.
- Sulman S, Rehman A. Isolation and Characterization of Cellulose Degrading Candida tropicalis W2 from Environmental Samples. Pakistan Journal of Zoology. 2013;45(3):809–816.
- Asliha I.N, Alami NH. Karakterisasi Khamir dari Pulau Poteran Madura [Characterization of yeast from Poteran Madura Island]. Jurnal Sains dan Seni ITS [Journal of Science and Art in ITS]. 2014;3(2):E49–E52. (In Indonesian). DOI: https://doi.org/10.12962/j23373520.v3i2.6869.
- Lopez S, Mateo JJ, Maicas S. Characterisation of Hanseniaspora Isolates with Potential Aroma-enhancing Properties in Muscat Wines. South African Journal of Enology and Viticulture. 2014;35(2):292–303.
- Mateo JJ, Peris L, Ibanez C, Maicas S. Characterization of glycolytic activities from non-Saccharomyces yeasts isolated from Bobal musts. Journal of Industrial Microbiology & Biotechnology. 2011;38(2):347–354. DOI: https://doi.org/10.1007/s10295-010-0780-z.
- Latouche GN, Daniel HM, Lee OC, Mitchell TG, Sorrell TC, Meyer W. Comparison of use of phenotypic and genotypic characteristics for identification of species of the anamorph genus Candida and related teleomorph yeast species. Journal of Clinical Microbiology. 1997;35(12):3171–3180.
- Shuler ML. Kargi F. Bioprocess Engineering: Basic Concepts, 2nd ed. Prentice Hall; 2001. 576 p.
- Nguong DLS, Jun LY, Yatim NI, Nathan S, Murad AMA, Mahadi NM, et al. Characterising Yeast Isolates from Malaysia towards the Development of Alternative Heterologous Protein Expression Systems. Sains Malaysiana. 2011;40(4):323–329.
- Ganzle MG, Ehmann M, Hammes WP. Modeling of growth of Lactobacillus sanfranciscensis and Candida milleri in response to process parameters of sourdough fermentation. Applied and Environmental Microbiology. 1998;64(7):2616–2623.
- Fennema OR. Food Chemistry, Third Edition. Taylor & Francis; 1996. 1067 p.
- Samaranayake YH, Samaranayake LP. Candida krusei: biology, epidemiology, pathogenicity and clinical manifestations of an emerging pathogen. Journal of Medical Microbiology. 1994;41(5):295–310. DOI: https://doi.org/10.1099/00222615-41-5-295.
- López S, Mateo JJ, Maicas S. Screening of Hanseniaspora Strains for the Production of Enzymes with Potential Interest for Winemaking. Fermentation. 2015;2(1). DOI: https://doi.org/10.3390/fermentation2010001.
- Lin Y, Zhang W, Li CJ, Sakakibara K, Tanaka S, Kong HN. Factors affecting ethanol fermentation using Saccharomyces cerevisiae BY4742. Biomass & Bioenergy. 2012;47:395–401. DOI: https://doi.org/10.1016/j.biombioe.2012.09.019.
- Putri F. Studies on Isolation, Identification and Characterization of Alcohol Fermentative Yeasts Indigenous to Vegetable and Fruit Waste in Indonesia. Japan: Mie University; 2018.
- Birmeta G, Bakeeva A, Passoth V. Yeasts and bacteria associated with kocho, an Ethiopian fermented food produced from enset (Ensete ventricosum). Antonie Van Leeuwenhoek International Journal of General and Molecular Microbiology. 2019;112(4):651–659. DOI: https://doi.org/10.1007/s10482-018-1192-8.
- Vervaeke S, Vandamme K, Boone E, De Laere E, Swinne D, Surmont I. A case of Candida lambica fungemia misidentified as Candida krusei in an intravenous drug abuser. Medical Mycology. 2008;46(8):853–856. DOI: https://doi.org/10.1080/13693780802342552.
- Harding MW, Butler N, Dmytriw W, Rajput S, Burke DA, Howard RJ. Characterization of Microorganisms from Fresh Produce in Alberta, Canada Reveals Novel Food-spoilage Fungi. Research Journal of Microbiology. 2017;12(1):20–32. DOI: http://doi.org/10.3923/jm.2017.20.32.
- Vontrobova E, Kubizniakova P, Fiala J, Sochor J, Matoulkova D. Autochthonous yeasts as one of the tools to produce wines by original technologies. Kvasny Prumysl. 2019;65(1):38–45. DOI: https://doi.org/10.18832/kp2019.65.38.
- Bengoa AA, Iraporda C, Garrote GL, Abraham AG. Kefir micro-organisms: their role in grain assembly and health properties of fermented milk. Journal of Applied Microbiology. 2019;126(3):686–700. DOI: https://doi.org/10.1111/jam. 14107.
- Miao YJ, Xiong GT, Li RY, Wu ZF, Zhang X, Weng PF. Transcriptome profiling of Issatchenkia orientalis under ethanol stress. Amb Express. 2018;8. DOI: https://doi.org/10.1186/s13568-018-0568-5.