Abstract
Phylogenetic information on microbial communities involved in fermenting botanicals has important implications for the food industry since it can provide a valuable perspective on the diversity, composition, and techno-functional properties and characteristics of the final product. Microbial phylogenetic analysis illustrates the evolutionary history of microbes through visual representational graphs (phylogenetic trees) showing the beginning and advancement of their assemblage.In this study, we used molecular methods to determine the phylogenetic identities of microbes occurring in spontaneously fermented sweet potato, maize, and pigeon pea samples after a 72-hourly evaluation every 12 h. The sequences obtained were edited using the bioinformatics algorithm against similar sequences downloaded from the National Center for Biotechnology Information (NCBI) database using BLASTN and aligned using ClustalX. The neighbor-joining technique was applied to extrapolate the chronicle of the isolates evolution.
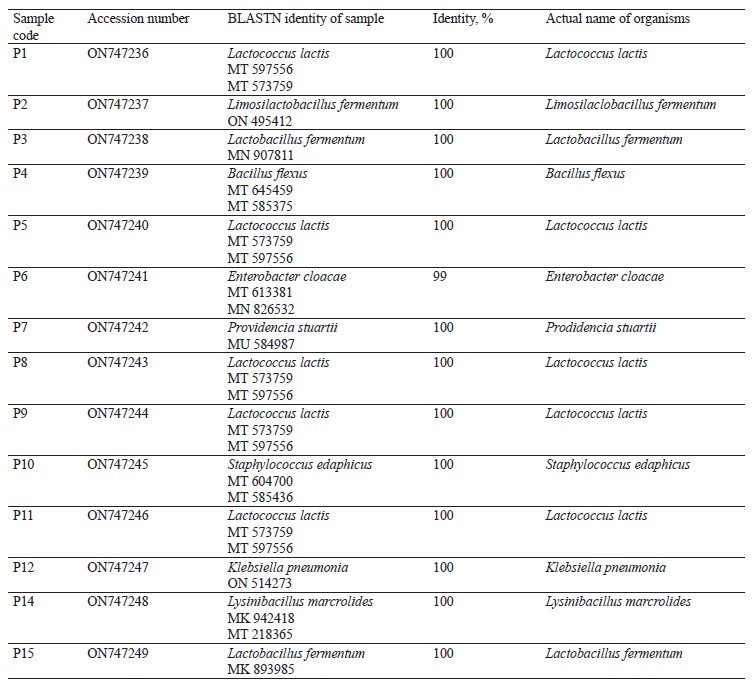
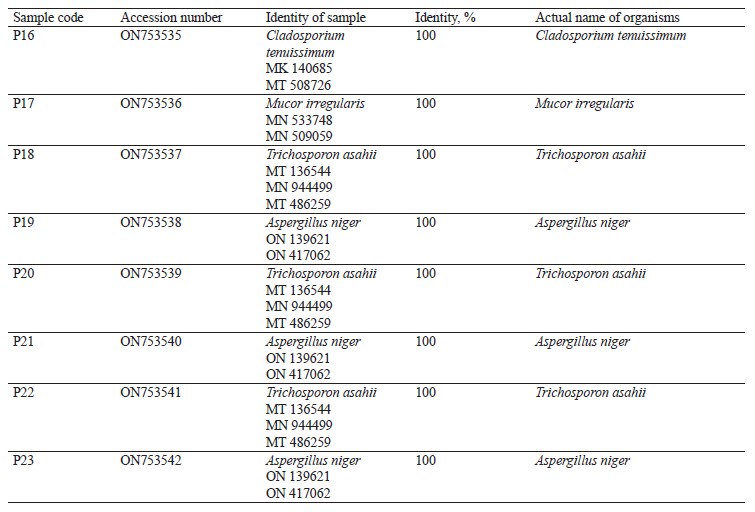
Molecular identification from the BLASTN results showed the following bacterial isolates: Lysinibacillus macrolides, Klebsiella pneumoniae, Lactococcus lactis, Providencia stuartii, Enterobacter cloacae, Limosilactobacillus fermentum, Lactobacillus fermentum, Staphylococcus edaphicus, and Bacillus flexus, as well as the following fungal isolates: Trichosporon asahii, Mucor irregularis, Cladosporium tenuissimum, and Aspergillus niger. The sequences obtained from the isolates produced an exact match with the NCBI non-redundant nucleotide (nr/nt) database. L. lactis had the highest percentage occurrence for bacteria (38.46%), while T. asahii and A. niger showed the highest occurrence for fungi (37.50%).
Identifying and characterizing the microorganisms involved in the fermentation process would allow optimizing fermentation conditions to enhance the quality and nutritional value of the final products.
Keywords
Phylogenetic identification, fermented botanicals, gluten-free composite, flour mixesINTRODUCTION
Microbial fermentation is a bio-engineered process that has been employed for centuries in the production of various nutrient-enhancing fermented food products such as beverages, bread, and dairy foods [1–5]. During this procedure, microorganisms such as bacteria and fungi enzymatically break down complex organic compounds inherent in the food substrate to produce various metabolites, including organic acids, alcohols, and gases [6]. Microbial communities involved in fermentation are diverse and complex, and their compositions and activities are influenced by various factors such as temperature, pH, and the presence of nutrients [7–9].
In recent years, there has been a growing interest in the use of botanicals as a substrate for microbial fermentation, particularly in the production of traditional fermented foods and beverages. The main purpose is to develop agro-processed, highly-nutritive, gluten-free therapeutic foods from comparatively advantageous indigenous botanicals [10–12]. Such botanicals include various plant materials (grains, vegetables, fruits, roots, and tubers) that are rich in nutrients and bioactive compounds and can influence the growth and metabolism of microorganisms [13]. Studies of microbial communities involved in the fermentation of botanicals have important implications for the food industry, as they can provide insights into the techno-functional properties and characteristics of the final product [12].
Customary methods for microbial characterization have been challenged overtime due to imprecise and ambiguous similarities they provide between different species [14]. This irregularity in morphological nomenclature for microorganisms necessitated a need for modern, more pragmatic and reliable taxonomic protocols. Phylogenetics is a molecular-based technique with an improved and more proficient method of characterizing and identifying microorganisms [15]. It entails studying progressive evolutionary relationships among organisms based on their genetic characteristics from a similar forebear [16]. This powerful tool for understanding the diversity and structure of microbial communities has become a ubiquitous part of biological analyses [17, 18].
Phylogenetic analysis of microbial isolates is one of the means by which one can learn about the evolutionary history of species by constructing comparative visual representational graphs of phylogenetic trees (illustrations showing the beginning and advancement of assemblage of organisms) using the organism’s morphological features [15, 19]. Recently, sequences from deoxyribonucleic acid (DNA) and proteins from different organisms have been used to determine their evolutionary relationships from common forebears to different off-springs [16, 20, 21].
Studying phylogenetic properties of microbial isolates is crucial in understanding the evolutionary history and diversity of microbial populations. Their evolutionary relationships provide insights into the origins, diversification, and distribution of microbial species. Phylogenetic properties can provide a better understanding of the microbial world, its interactions with the environment, and its impact on human health and disease [22]. This knowledge can be applied in various fields, such as biotechnology, medicine, and agriculture, to develop innovative solutions to tackle the challenges we face today [23].
In this study, we aimed to phylogenetically identify microorganisms isolated from fermented botanicals used to formulate gluten-free composite flour mixes.
STUDY OBJECTS AND METHODS
Collection and confirmation of samples. Yellowfleshed sweet potato tubers (Ipomea batatas L.), maize of the yellow-grain variety (Zea mays L.), and pigeon peas (Cajanus cajan L. Millsp.) were obtained from local food merchants in the Auchi metropolis, specifically in the Etsako-West Local Government Area of Edo State, Nigeria. The authenticity and quality of these samples were verified at the Herbarium Curation Division, Department of Basic Sciences, Edo University Uzairue, also located in Edo State, Nigeria.
Preparation and fermentation of samples. The samples of botanical materials were subjected to fermentation, which was carried out spontaneously for 72 h. The process took place at 28 ± 2°C, following the procedures described in [12].
Microbiological analysis. The fermented botanical samples underwent microbiological analysis every 12 h to determine the total microbial counts. The method used for this analysis followed the guidelines provided by the American Public Health Association [24, 25]. To initiate the analysis, 1 mL of the fermented samples was aseptically withdrawn and mixed with 9 mL of peptone water. Subsequently, we performed a sequential 10-fold dilution.
For the microbiological analysis, aliquots from the final dilutions were taken and introduced into specific agar media. Bacteria were cultured using Nutrient Agar (NA), MacConkey Agar (MCA), and De Mann-Rogosa-Sharpe Agar (MRS). Fungi, on the other hand, were cultured using Potato Dextrose Agar (PDA). The plates containing the samples and agar media were then incubated for 24–48 h at 37°C for bacteria and at room temperature (25 ± 2°C) for fungi.
All of these processes, including dilution, culturing, and incubation, were conducted in triplicate to ensure accuracy and reliability of the results.
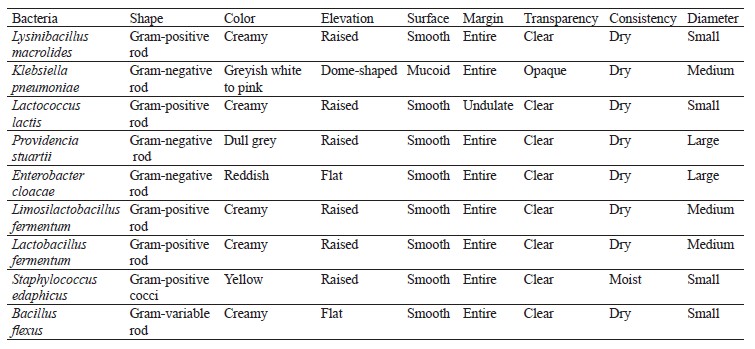
Isolation and enumeration of bacteria and fungi. Distinct colonies with varying morphologies were counted and reported as colony-forming units per milliliter (CFU/mL) of the respective samples. To classify these colonies, they were separated as pure cultures and preserved in agar slants both at 4°C and at room temperature. Standard morphological, biochemical, and molecular techniques were employed to confirm the identification of the different bacterial and fungal species [26, 27].
Molecular identification. Bacterial genomic DNA extraction. Five milliliters of an overnight liquid culture of the bacterial isolate in Luria Bertani (LB) medium was centrifuged at 14 000 rpm for 3 min. The resulting pellet was then resuspended in 500 mL of normal saline and heated at 95°C for 20 min. After cooling on ice, the mixture was spun for another 3 min at 14 000 rpm. The resulting supernatant, which contained the DNA, was carefully removed and transferred to a 1.5-mL micro centrifuge tube. This DNA extract was then stored at –20°C for future downstream reactions.
DNA quantification. The genomic DNA obtained was measured for its quantity using a Nanodrop 1000 spectrophotometer. To initiate the process, the Nanodrop software was opened by double-clicking on the Nanodrop icon. The spectrophotometer was calibrated using 2 µL of sterile distilled water as the initial blank, which was replaced with normal saline. Next, 2 µL of the extracted DNA was carefully loaded onto the lower pedestal of the spectrophotometer, and the upper pedestal was lowered to allow contact with the DNA sample. When the “measure” button was clicked, the Nanodrop spectrophotometer provided the measurement of the DNA concentration.
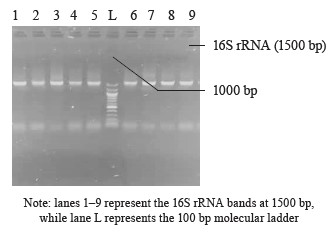
16S rRNA amplification. The 16S ribosomal RNA (rRNA) region of the rRNA genes in the isolates was amplified using the 27F (5’-AGAGTTTGATCMTGGCTCAG-3’) and 1492R (5’-CGGTTACCTTGTTACGACTT-3’) primers. The amplification process was carried out in a 50 µL final volume for 35 cycles, using an ABI 9700 Applied Biosystems thermal cycler. The PCR mix consisted of the X2 Dream Taq Master Mix provided by Inqaba, South Africa, which included taq polymerase, DNTPs, and MgCl, along with the primers at a concentration of 0.4 M. The extracted DNA served as a template for the PCR reaction. The PCR reaction conditions were as follows: initial denaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 52°C for 30 s, extension at 72°C for 30 s, which was repeated for 35 cycles, and final extension at 72°C for 5 min. The resulting PCR product was separated on 1% agarose gel, subjected to electrophoresis at 120 V for 15 min, and visualized using a UV transilluminator.
Fungal genomic DNA extraction. DNA extraction was performed using a ZR fungal DNA mini prep extraction kit provided by Inqaba, South Africa. Fungal isolates in pure culture were densely grown and suspended in 200 µL of isotonic buffer, to be then transferred to ZR Bashing Bead Lysis tubes. To this, 750 µL of lysis solution was added, and the tubes were placed securely in a bead beater equipped with a 2-mL tube holder assembly. The samples underwent processing at maximum speed for 5 min.
Subsequently, the ZR Bashing Bead Lysis tubes were centrifuged at 10 000 × g for 1 min. After centrifugation, 400 µL of the supernatant was carefully transferred to a Zymo-Spin IV spin Filter (orange top) positioned in a collection tube. The collection tube was then centrifuged at 7000 × g per 1 min. Then, 1200 µL of fungal/bacterial DNA binding buffer was added to the filtered liquid in the collection tube, resulting in a final volume of 1600 µL. Next, 800 µL of this mixture was moved to a Zymo-Spin IIC column placed in the collection tube, which was centrifuged at 10 000 × g for 1 min. The flowthrough was discarded, and the remaining volume was retained within the Zymo-Spin IIC column.
Then, 200 µL of DNA pre-wash buffer was added to the Zymo-Spin IIC column in a fresh collection tube and centrifuged at 10 000 × g for 1 min. Following this, 500 µL of fungal/bacterial DNA wash buffer was added to the column, and centrifugation was performed at 10 000 × g for 1 min. The Zymo-Spin IIC column was then transferred to a clean 1.5-µL centrifuge tube. To elute the DNA, 100 µL of DNA elution buffer was added to the column matrix and centrifuged at 10 000 × g for 30 s. The resulting DNA, which was of high purity, was stored at –20°C for subsequent downstream reactions.
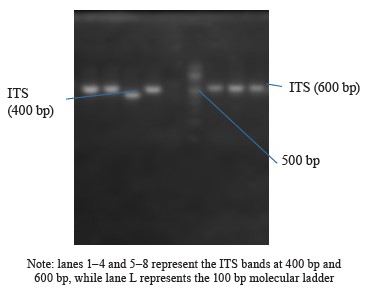
Internal Transcribed Spacer (ITS) amplification. The ITS region of the rRNA genes present in the fungal isolates was amplified using specific primers, namely ITS1-F: 5’-TCCGTAGGTGAACCTGCGG-3’ and ITS4-R: 5’-TCCTCCGCTTATTGATATGC-3’. This amplification process was carried out on an ABI 9700 Applied Biosystems thermal cycler, with a final reaction volume of 50 µL for a total of 35 cycles. The PCR mixture consisted of the X2 Dream Taq Master Mix provided by Inqaba, South Africa, which contained taq polymerase, DNTPs, and MgCl, along with the primers at a concentration of 0.4 M. The extracted DNA served as a template for the PCR reaction. The PCR conditions involved initial denaturation at 95°C for 5 min, followed by denaturation at 95°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 30 s. These steps were repeated for 35 cycles, and a final extension was performed at 72°C for 5 min. To visualize the PCR product, it was separated on a 1% agarose gel using an electric field of 120V for 15 min, and the resulting bands were observed under a UV transilluminator.
Sequencing was conducted at Inqaba Biotechnological in Pretoria, South Africa, using the BigDye Terminator Kit on a 3510 ABI sequencer. The sequencing reaction was prepared with a final volume of 10 µL, consisting of the following components: 0.25 µL of BigDye® Terminator v1.1/v3.1, 2.25 µL of 5× BigDye sequencing buffer, 10 micromolar Primer PCR primer, and 2–10 nanograms of PCR template per 100 base pairs. The sequencing process involved 32 cycles with the following temperature conditions: denaturation at 96°C for 10 s, annealing at 55°C for 5 s, and extension at 60°C for 4 min.
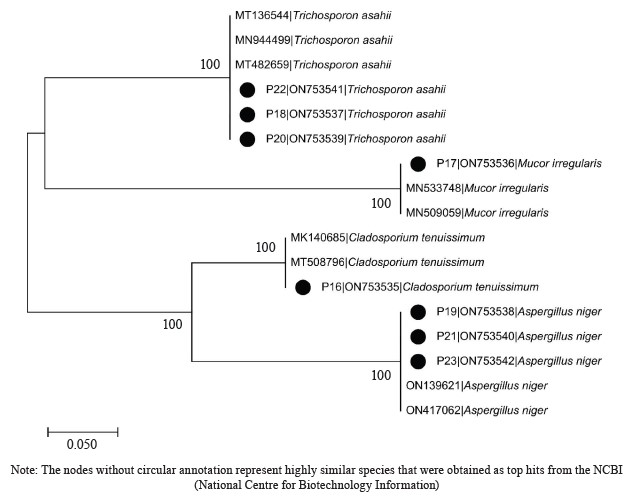
Phylogenetic analysis of the isolates. The obtained sequences underwent editing using the bioinformatics algorithm Trace Edit. To identify similar sequences, we used the National Center for Biotechnology Information (NCBI) database. These sequences were then aligned using ClustalX. The evolutionary history of the isolates was inferred using the neighbor-joining technique [28]. The resulting phylogenetic tree displayed the most supported relationships, with a branch length summation of 0.73390024. To assess the reliability of the tree, a bootstrap test with 1000 replicates was conducted, showing the percentage of replicate trees in which the taxa were grouped together near the branches [29]. The tree was visualized with branch lengths reflecting the inferred evolutionary distances. The phylogenetic analysis involved the computation of the phylogenetic space using the Jukes-Cantor procedure, considering 17 nucleotide progressions [30]. Sites with less than 95% inclusion were removed, representing less than 5% of the sequence space, and positions allowing for cryptic bases were retained using a partial removal selection approach. The final dataset consisted of 324 positions. The evolutionary analysis was performed using MEGA X [31].
RESULTS AND DISCUSSION
Molecular identification from the BLASTN results of the DNA sequences shows that the 16S rRNA sequence obtained from the isolate produced an exact match during the megablast search for highly similar sequences from the NCBI non-redundant nucleotide (nr/nt) database (Fig. 1). The 16S rRNA of the isolates W1 showed the 99–100 % percentage similarity to other species. The evolutionary distances computed using the Jukes-Cantor method were in agreement with the phylogenetic placement of the 16S rRNA of the isolates within the Providencia, Enterobacter, Klebsiella, Lysinibacillus, Staphylococcus, Limosilactobacillus, Lactobacillus, and Lactococcus sp. They also revealed a close relatedness to Providencia stuartii, Enterobacter cloacae, Klebsiella pneumoniae, Lysinibacillus macroides, Staphylococcus edaphicus, Limosilactobacillus fermentum, Lactobacillus fermentum, and Lactococcus lactis (Fig. 2).
The DNA sequencing results using BLASTN identified the following fungi isolates (Fig. 4).
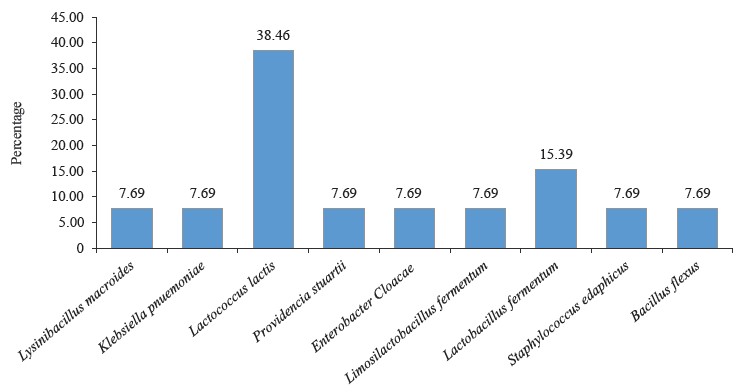
L. lactis and L. fermentum were the highest occurring bacteria isolates with percentage occurrences of 38.46 and 15.39%, respectively, while L. fermentum occurred least with a 7.69% occurrence (Fig. 5). Lactic acid bacteria are a phylogenetically heterogeneous group of Gram-positive bacteria that share metabolic and physiological characteristics [32]. These bacteria ferment carbohydrates into lactic acid (homofermentative), or into lactic acid, ethanol, and CO2 (heterofermentative) [33, 34].
L. lactis is widely used in industry as a fast acidifier, starter culture, and flavor enhancer in milk fermentation [35–37]. In addition, L. lactis subsp. lactis is widely applied in cheese and butter production, as well as to inhibit pathogen development [38–41].
Food-associated Lactobacillus strains are “generally recognized as safe” (GRAS) microorganisms that have a major role in fermented milk production and in sourdough technology. In particular, some members of these strains called non-starter lactic cultures play a considerable role in developing cheese and dough aroma, texture, and flavor through ripening. They are currently being used in fortifying health-enhancing functional foods [42–44].
L. fermentum is a Gram-positive, non-spore-forming, rod-shaped probiotic bacterium belonging to the heterofermentative lactic acid bacterium. It is capable of fermenting carbohydrates and producing lactic and organic acids [45]. The bacterium has anti-diabetic probiotic features which enable host cells to adjust their anti-inflammatory and antioxidant systems, resulting in improved glucose homeostasis capable of oxidative stress protection in diabetic conditions [46, 47]. It can be added to fermented foods like yoghurt and is found in some dietary supplements [48, 49]. In addition, L. fermentum was reported as the most predominant bacteria in Chinese cereal gruel, West African cereal dough, and Indian rice-based fermented beverage [50–52]. Furthermore, it is regarded as “generally recognized as safe” (GRAS) by the United States Food and Drug Administration (FDA) [53]. L. fermentum can also inhibit the growth of foodborne pathogens in food products [54]. In addition, foods obtained from fermentation by L. fermentum usually possess good palatability, high sensory quality, texture, stability, and nutritional properties [55, 56].
The spore-forming bacteria include L. macroides and Bacillus flexus with an occurrence of 7.69% (Fig. 5). They demonstrate an exceptional ability to adapt to their environment, and their presence in fermented flours could be due to their capacity to resist the acid produced by lactic acid bacteria [55–59]. Also, they are commonly found in beans and legumes [60]. Bacillus spp. present in fermented foods hydrolyze the substrate and produce enzymes, such as nattokinase, phytase, amylase, protease, cellulase, and lipase. These enzymes help break down complex compounds into simple biomolecules [61]. For example, amylase converts starch in legumes into sugar. Likewise, protease is used to convert proteins into amino acids [61, 62].
Bacillus species have also shown probiotic potential [63]. B. flexus biofilm has been used as a biological control agent against the Cowpea pest Callosobruchus maculates [64].The microbial profile of the fermented flour also showed the presence of Enterobacteriaceae. These include Klebsiella pnuemoniae, P. stuarti, and E. cloacae with a 7.69% occurrence each (Fig. 5). The Enterobacteriaceae family comprises a large group of Gram-negative nonspore-forming bacteria. These facultative anaerobic rods, which break down glucose-producing acid with/without gas, include some harmless commensal species, as well as important human and animal pathogens [65]. Their ubiquitous distribution means some members of the Enterobacteriaceae family will inevitably enter the food chain. While their low numbers are acceptable and do not directly lead to safety apprehension, their presence signifies inappropriate or poor processing and sanitary protocols around the food processing surroundings [66, 67].
Generally, Enterobacteriaceae are considered hygiene indicator organisms during food processing. Therefore, they are used to monitor the effectiveness of implemented preventive pre-requisite measures such as Good Manufacturing Practices and Good Hygiene Practices (GMP/GHP) [68].
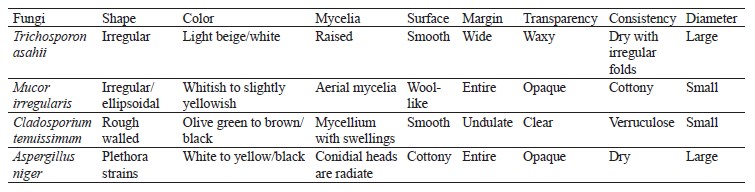
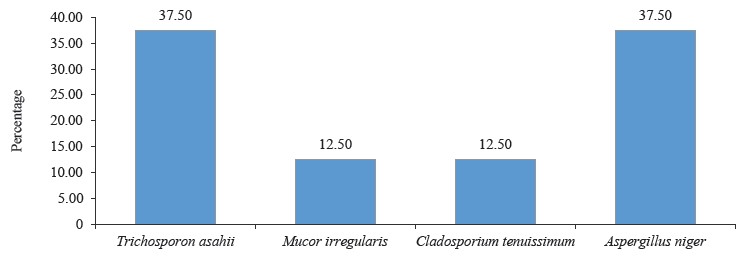
According to percentage occurrence data for the fungi from fermented botanicals, both T. asahii and A. niger had a 37.50% occurrence, while M. irregularis and Cladosporium temuissimum had an occurrence of 12.50% (Fig. 6). These organisms are commonly present as contaminants in the human skin, cooking utensils, processing equipment, the environment, water, or in the seeds of cereals and legumes [69, 70]. They do not appear to play a significant role in the fermentation process, although they could be further exploited for their probiotic potentials [70–72].
CONCLUSION
The amalgamation of morphological attributes and molecular (DNA) markers have been accurately used for microbial nomenclature at the molecular level because the processes eliciting genetic changeability are the direct product of sequence changes from biochemical markers (genes and proteins). The phylogenetic properties of microbial isolates obtained from the fermentation of botanicals can provide valuable information on the diversity, composition, and functional properties of these microbial communities. Identification and characterization of the microorganisms involved in the fermentation process may optimize fermentation conditions to enhance the quality and nutritional value of the final products.
Contribution
All the authors participated in developing the research concept and writing the original draft.
CONFLICTS OF INTEREST
The authors have no conflict of interest concerning the conceptualization, research design, and publication of this work.
ACKNOWLEDGEMENTS
All the data associated with this study has been deposited in the NCBI GenBank database, with the accession numbers of ON747236-ON747249 (bacteria) and ON753535-ON753542 (fungi).
FUNDING
This study was funded entirely by Auchi Polytechnic , Auchi, Edo State, Nigeria through the Nigerian Tertiary Education Trust Fund (TETFUND) under the reference number TETF/DR&D/CE/POLY/AUCHI/IBR/2022/VOL.I/BATCH 10.REFERENCES
- Marco ML, Heeney D, Binda S, Cifelli CJ, Cotter PD, Foligné B, et al. Health benefits of fermented foods: microbiota and beyond. Current Opinion in Biotechnology 2017;44:94–102. https://doi.org/10.1016/j.copbio.2016.11.010
- Llorente B, Williams TC, Goold HD, Pretorius SI, Paulsen IT. Harnessing bioengineered microbes as a versatile platform for space nutrition. Nature Communications. 2022;13. https://doi.org/10.1038/s41467-022-33974-7
- Plessas S. the rendering of traditional fermented foods in human diet: Distribution of health benefits and nutritional benefits. Fermentation 2022;8(12). https://doi.org/10.3390/fermentation8120751
- Krikunova LN, Meleshkina EP, Vitol IS, Dubinina EV, Obodeeva ON. Grain bran hydrolysates in the production of fruit distillates. Foods and Raw Materials. 2023;11(1):35–42. https://doi.org/10.21603/2308-4057-2023-1-550
- Gryaznova MV, Burakova IYu, Smirnova YuD, Nesterova EYu, Rodionova NS, Popov ES, et al. Bacterial composition of dairy base during fermentation. Food Processing: Techniques and Technology. 2023;53(3):554–564. (In Russ.). https://doi.org/10.21603/2074-9414-2023-3-2456
- Taveira IC, Nogueira KMV, de Oliveira DLG, Silva RN. Fermentation: Humanity’s oldest biotechnological tool. Frontiers for Young Minds. 2021; 9. https://doi.org/10.3389/frym.2021.568656
- Sanlier N, Gökcen BB, Sezgin AC. Health benefits of fermented foods. Critical Reviews in Food Science and Nutrition. 2019;59(3):506–527. https://doi.org/10.1080/10408398.2017.1383355
- Sharma R, Garg P, Kumar P, Bhatia SK, Kulshrestha S. Microbial fermentation and its role in quality improvement of fermented foods. Fermentation. 2020;6(4). https://doi.org/10.3390/fermentation6040106
- Voidarou C, Antoniadou M, Rozos G, Tzora A, Skoufos I, Varzakas T, et al. Fermentative foods: Microbiology, biochemistry, potential human health benefits and public health issues. Foods. 2021;10(1). https://doi.org/10.3390/foods10010069
- Kumari M, Platel K. Impact of soaking, germination, fermentation, and thermal processing on the bioaccessibility of trace minerals from food grains. Journal of Food Processing and Preservation. 2020;44(10). https://doi.org/10.1111/jfpp.14752
- Galanakis CM. Functionality of food components and emerging technologies. Foods. 2021;10(1). https://doi.org/10.3390/foods10010128
- Oleghe PO, Oladebeye AA, Johnson DO. Microbiological and techno-functional assessment of unfermented and fermented gluten-free flour mixes. International Journal of Life Sciences Research. 2023;11(1):33–48. https://doi.org/10.5281/zenodo.7702091
- Chan M, Liu D, Wu Y, Yang F, Howell K. Microorganisms in whole botanical fermented foods survive processing and simulated digestion to affect gut microbiota composition. Frontiers of Microbiolology. 2021;12. https://doi.org/10.3389/fmicb.2021.759708
- Som A. Causes, consequences and solutions of phylogenetic incongruence. Briefings in Bioinformatics. 2015;16(3):536–548. https://doi.org/10.1093/bib/bbu015
- Adebayo EA, Elkanah FA, Afolabi FJ, Ogundun OS, Alabi TF, Oduoye OT. Molecular characterization of most cultivated Pleurotus species in sub-western region Nigeria with development of cost effective cultivation protocol on palm oil waste. Heliyon. 2021;7(2). https://doi.org/10.1016/j.heliyon.2021.e06215
- Kaari M, Joseph J, Manikkam R, Shamya M, Aruni W. Appliication of bioinformatic tools for phylogenetic analysis. In: Dharumadurai D, editor. Methods in actinobacteriology. New York: Humana; 2022. pp. 187–191. https://doi.org/10.1007/978-1-0716-1728-1_27
- Bawono P, Heringa J. Phylogenetic analyses. In: Brahme A, editor. Comprehensive biomedical physics. Vol. 6. Elsevier; 2014. pp. 93–110. https://doi.org/10.1016/B978-0-444-53632-7.01108-4
- Wang X, Weber GF. Quantitative analysis of protein evolution: The phylogeny of osteopontin. Frontiers in Genetics. 2021;12. https://doi.org/10.3389/fgene.2021.700789
- Mclennan DA. How to read a phylogenetic tree. Evolution: Education and Outreach. 2010;3:506–519. https://doi.org/10.1007/s12052-010-0273-6
- Baum D. Reading a phylogenetic tree: The meaning of monophyletic groups. Nature Education. 2008;1(1).
- Weber GF. The phylogeny of Osteopontin – Analysis of the protein sequence. International Journal of Molecular Sciences. 2018;19(9). https://doi.org/10.3390/ijms19092557
- Podsiadlo L, Polz-Dacewicz M. Molecular evolution and phylogenetic implications in clinical research. Annals of Agricultural and Environmental Medicine. 2013;20(3):455–459.
- Abzhanov A. Phylogenetic analysis and it’s applications. Journal of Phylogenetics and Evolutionary Biology. 2021;9(8).
- Compendium of methods for the microbiological examination of foods. 4th edition. Washington: American Public Health Association; 2001. 676 p.
- Oleghe PO, Orhewere RDA, Orhewere VA, Oboh JE. Microwave heat treatment effects on the microbial profile of some ready-to-eat street vended snacks. International Journal of Scientific Research in Biological Sciences. 2022;9(2):76–83. https://doi.org/10.26438/ijsrbs/v9i2.7683
- Abdalla MOM, Omer HEA. Microbiological characteristics of white cheese (Gibna bayda) manufactured under traditional conditions. Journal of Advances in Microbiology. 2017;2(3):1–7. https://doi.org/10.9734/jamb
- Alsohaili SA, Bani-Hasan BM. Morphological and molecular identification of fungi isolated from different environmental sources in the northern eastern Desert of Jordan. Jordan Journal of Biological Sciences. 2018;11(3):329–337.
- Saitou N, Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39(4):783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. Volume III. Academic Press; 1969. pp. 21–132. https://doi.org/10.1016/B978-1-4832-3211-9.50009-7
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution. 2018;35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
- Ganzle MG. Lactic metabolism revisited: Metabolism of lactic acid bacteria in food fermentations and food spoilage. Current Opinion in Food Science. 2015;2:106–117. https://doi.org/10.1016/j.cofs.2015.03.001
- Bintsis T. Lactic acid bacteria: Their applications in foods. Journal of Bacteriology and Mycology: Open Access. 2018;6(2):89–94. https://doi.org/10.15406/jbmoa.2018.06.00182
- Ayivi RD, Gyawali R, Krastanov A, Aljaloud SO, Worku M, Tahergorabi R, et al. Lactic acid bacteria: Food safety and human health applications. Dairy. 2020;1(3):202–232. https://doi.org/10.3390/dairy1030015
- Omaea M, Maeyama Y, Nishimura T. Sensory properties and taste compounds of fermented milk produced by Lactococcus lactis and Streptococcus thermophilus. Food Science and Technology Research. 2008;14(2):183–189. https://doi.org/10.3136/fstr.14.183
- Cavanagh D, Fitzgerald GF, McAuliffe O. From field to fermentation: The origins of Lactococcus lactis and its domestication to the dairy environment. Food Microbiology 2015;47:45–61. https://doi.org/10.1016/j.fm.2014.11.001
- Li W, Ren M, Duo L, Li J, Wang S, Sun Y, et al. Fermentation characteristics of Lactococcus lactis subsp. lactis isolated from naturally fermented dairy products and screening of potential starter isolates. Frontiers in Microbiology. 2020;11. https://doi.org/10.3389/fmicb.2020.01794
- Madera C, García P, Janzen T, Rodríguez A, Suárez JE. Characterisation of technologically proficient wild Lactococcus lactis strains resistant to phage infection. International Journal of Food Microbiology. 2003;86(3):213–222. https://doi.org/10.1016/s0168-1605(03)00042-4
- Özkalp B, Özden B, Tuncer Y, Sanlibaba P, Akçelik, M. Technological characterization of wild-type Lactococcus lactis strains isolated from raw milk and traditional fermented milk products in Turkey. Le Lait. 2007;87(6):521–534. https://doi.org/10.1051/lait:2007033
- Kleerebezem M, Bachmann H, van Pelt-KleinJan E, Douwenga S, Smid EJ, Teusink B, et al. Lifestyle, metabolism and environmental adaptation in Lactococcus lactis. FEMS Microbiology Reviews. 2020;44(6):804–820. https://doi.org/10.1093/femsre/fuaa033
- Maalaoui A, Trimeche A, Marnet PG, Demarigny Y. Use of Lactococcus lactis subsp. Lactis strains to inhibit the development of pathogens. Food and Nutrition Sciences. 2020;11(2):98–112. https://doi.org/10.4236/fns.2020.112009
- Aziz K, Haseeb-Zaidi A, Fatima HN, Tariq M. Lactobacillus fermentum strains of dairy-product origin adhere to mucin and survive digestive juices. Journal of Medical Microbiology. 2019;68(12):1771–1786. https://doi.org/10.1099/jmm.0.001090
- Naghmouchi K, Belguesmia Y, Bendali F, Spano G, Seal BS, Drider D. Lactobacillus fermentum: A bacterial species with potential for food preservation and biomedical applications. Critical Review in Food Science and Nutrition. 2020;60(20):3387–3399. https://doi.org/10.1080/10408398.2019.1688250
- Bobga PT, Fossi BT, Taiwe GS, Nkanpira KT, Yolande NE, Ngwa FA, et al. Evaluation of the anti-diabetic potential of probiotic Lactobacillus fermentum (PRI 29) isolated from Cameroonian fermented cow milk in alloxan induced diabetes type-1 mice model. Saudi Journal of Pathology and Microbiology. 2022;7(10):381–393. https://doi.org/10.36348/sjpm.2022.v07i10.001
- Wai SN, How YH, Saleena LAK, Degraeve P, Oulahal N, Pui LP. Chitosan–sodium caseinate composite edible film incorporated with probiotic Limosilactobacillus fermentum: Physical properties, viability, and antibacterial properties. Foods. 2022;11(22). https://doi.org/10.3390/foods11223583
- Lacerda DC, Trindade da Costa PC, Pontes PB, Carneiro dos Santos LA, Cruz Neto JPR, et al. Potential role of Limosilactobacillus fermentum as a probiotic with antidiabetic properties: A review. World Journal of Diabetes. 2022;13(9):717–728. https://doi.org/10.4239/wjd.v13.i9.717
- Paulino do Nascimento LC, Lacerda DC, Ferreira DJS, de Souza EL, de Brito Alves JL. Limosilactobacillus fermentum, current evidence on the antioxidant properties and opportunities to be exploited as a probiotic microorganism. Probiotics and Antimicrobial Proteins. 2022;14:960–979. https://doi.org/10.1007/s12602-022-09943-3
- D’ambrosio S, Ventrone M, Fusco A, Casillo A, Dabous A, Cammarota M, et al. Limosilactobacillus fermentum from buffalo milk is suitable for potential biotechnological process development and inhibits Helicobacter pylori in a gastric epithelial cell model. Biotechnology Reports. 2022;34. https://doi.org/10.1016/j.btre.2022.e00732
- Kawai T, Ohshima T, Tanaka T, Ikawa S, Tani A, Inazumi N, et al. Limosilactobacillus (Lactobacillus) fermentum ALAL020, a probiotic candidate bacterium, produces a cyclic dipeptide that suppresses the periodontal pathogens Porphyromonas gingivalis and Prevotella intermedia. Frontiers in Cellular and Infection Microbiology. 2022;12. https://doi.org/10.3389/fcimb.2022.804334
- Qin H, Sun Q, Pan X, Qiao Z, Yang H. Microbial diversity and biochemical analysis of Suanzhou: A traditional Chinese fermented cereal gruel. Frontiers in Microbiology. 2016;7. https://doi.org/10.3389/fmicb.2016.01311
- Houngbédji M, Johansen P, Padonou SW, Akissoé N, Arneborg N, Nielsen DS, et al. Occurrence of lactic acid bacteria and yeasts at species and strain level during spontaneous fermentation of mawè, a cereal dough produced in West Africa. Food Microbiology. 2018;76:267–278. https://doi.org/10.1016/j.fm.2018.06.005
- Ghosh K, Ray M, Adak A, Halder SK, Das A, Jana A, et al. Role of probiotic Lactobacillus fermentum KKL1 in the preparation of a rice based fermented beverage. Bioresource Technology. 2015;188:161–168. https://doi.org/10.1016/j.biortech.2015.01.130
- Roberts A, Haighton LA. A hard look at FDA’s review of GRAS notices. Regulatory Toxicology and Pharmacology. 2016;79(S2):S124–S128. https://doi.org/10.1016/j.yrtph.2016.06.011
- Hossain TJ, Mozumder HA, Ali F, Akther K. Inhibition of pathogenic microbes by the lactic acid bacteria Limosilactobacillus fermentum strain LAB-1 and Levilactobacillus brevis strain LAB-5 isolated from the dairy beverage borhani. Current Research in Nutrition and Food Science. 2022;10(3):928–939. https://doi.org/10.12944/CRNFSJ.10.3.10
- Ale EC, Rojas MF, Reinheimer JA, Binetti AG. Lactobacillus fermentum: Could EPS production ability be responsible for functional properties? Food Microbiology. 2020;90. https://doi.org/10.1016/j.fm.2020.103465
- Pakroo S, Tarrah A, Takur R, Wu M, Corich V, Giacomini A. Limosilactobacillus fermentum ING8, a potential multifunctional non-starter strain with relevant technological properties and antimicrobial activity. Foods. 2022;11(5). https://doi.org/10.3390/foods11050703
- Aramesh M, Ajoudanifar H. Alkaline protease producing Bacillus isolation and identification from Iran. Banat's Journal of Biotechnology. 2017;8(6):140–147.
- Zhu X, Sun T, Sun X, Chen H, He H, Duan H, et al. Lysinibacillus macroides 38328, a potential probiotics strain, enhances antioxidant capacity and avian influenza virus vaccine immune response in laying hens. https://doi.org/10.21203/rs.3.rs-2262947/v1
- Chen H, Sun X, He H, Hong R, Duan H, Zhang C, et al. Lysinibacillus macrolides 38328 isolated from agricultural soils as a promising probiotic candidate for intestinal health. https://doi.org/10.21203/rs.3.rs-1881088/v1
- Tilak KVBR, Ranganayaki N, Pal KK, De R, Saxena AK, Shekhar-Nautiyal C, et al. Diversity of plant growth and soil health supporting bacteria. Current Science. 2005; 89(1):136–150.
- Gopikrishna T, Suresh-Kumar HK, Perumal K, Elangovan E. Impact of Bacillus in fermented soybean foods on human health. Annals of Microbiology. 2021;71. https://doi.org/10.1186/s13213-021-01641-9
- Rai AK, Sanjukta S, Chourasia R, Bhat I, Bhardwaj PK, Sahoo D. Production of bioactive hydrolysate using protease, β-glucosidase and α-amylase of Bacillus spp. isolated from kinema. Bioresource Technology. 2017;235:358–365. https://doi.org/10.1016/j.biortech.2017.03.139
- Gayathri L, Krubha A. Bacillus species–Elucidating the dilemma on their probiotic and pathogenic traits. In: Dhanasekaran D, Sankaranarayanan A, editors. Advances in probiotics: Microorganisms in food and health. Academic Press; 2021. pp 233–245. https://doi.org/10.1016/B978-0-12-822909-5.00015-0
- Reda FM, Hassanein WA, Moabed S, El-Shafiey SN. Potential exploitation of Bacillus flexus biofilm against the cowpea weevil, Callosobruchus maculates (F.) (Coleoptera: Bruchidae). Egyptian Journal of Biological Pest Control. 2020;30. https://doi.org/10.1186/s41938-020-00222-3
- Nelson GE, Greene MH. Enterobacteriaceae: In: Bennett JE, Dolin R, Blaser MJ, editors. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. Elsevier; 2020. pp. 2669–2685.
- Baylis CL. Enterobacteriaceae. In: Blackburn CW, editor. Food spoilage microorganisms. Woodhead Publishing; 2006. pp. 624–667. https://doi.org/10.1533/9781845691417.5.624
- Li P, Jiang H, Xiong J, Fu M, Huang X, Huang B, et al. Foodborne pathogens of enterobacteriaceae, their detection and control. In: Bhardwaj SB, editor. Entrobacteria. IntechOpen; 2022. https://doi.org/10.5772/intechopen.102086
- Malavi DN, Muzhingi T, Abong GO. Good manufacturing practices and microbial contamination sources in orange fleshed sweet potato puree processing plant in Kenya. International Journal of Food Science. 2018;2018. https://doi.org/10.1155/2018/4093161
- Bockelmann W. Cheese. Smear-ripened cheeses. In: Fuquay JW, editor. Encyclopedia of dairy sciences. Academic Press; 2011. pp. 753–766. https://doi.org/10.1016/B978-0-12-374407-4.00089-3
- Omemu AM, Okafor UI, Obadina AO, Bankole MO, Adeyeye SOA. Microbiological assessment of Maize ogi cofermented with pigeon pea. Food Science and Nutrition. 2018;6(5):1238–1253. https://doi.org/10.1002/fsn3.651
- Mbata TI, Ikenebomeh MJ, Alaneme JC. Studies on the microbiological, nutrient composition and antinutritional contents of fermented maize flour fortified with bambara groundnut (Vigna subterranean L). African Journal of Food science. 2009;3(6):165–171.
- Shruthi B, Deepa N, Somashekaraiah R, Adithi G, Divyashree S, Sreenivasa MY. Exploring biotechnological and functional characteristics of probiotic yeasts: A review. Biotechnology Reports. 2022;34. https://doi.org/10.1016/j.btre.2022.e00716